Evolutionary shift detection with ensemble variable selection
- PMID: 38245667
- PMCID: PMC10800078
- DOI: 10.1186/s12862-024-02201-w
Evolutionary shift detection with ensemble variable selection
Abstract
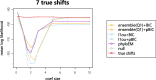
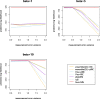
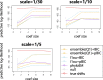
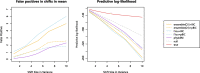
Abrupt environmental changes can lead to evolutionary shifts in trait evolution. Identifying these shifts is an important step in understanding the evolutionary history of phenotypes. The detection performances of different methods are influenced by many factors, including different numbers of shifts, shift sizes, where a shift occurs on a tree, and the types of phylogenetic structure. Furthermore, the model assumptions are oversimplified, so are likely to be violated in real data, which could cause the methods to fail. We perform simulations to assess the effect of these factors on the performance of shift detection methods. To make the comparisons more complete, we also propose an ensemble variable selection method (R package ELPASO) and compare it with existing methods (R packages [Formula: see text]1ou and PhylogeneticEM). The performances of methods are highly dependent on the selection criterion. [Formula: see text]1ou+pBIC is usually the most conservative method and it performs well when signal sizes are large. [Formula: see text]1ou+BIC is the least conservative method and it performs well when signal sizes are small. The ensemble method provides more balanced choices between those two methods. Moreover, the performances of all methods are heavily impacted by measurement error, tree reconstruction error and shifts in variance.
Keywords: ELPASO; Ensemble method; Evolutionary shift detection; LASSO; Ornstein-Uhlenbeck model; Phylogenetic comparative methods; Trait evolution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













References
-
- Felsenstein J. Phylogenies and the Comparative Method. Am Nat. 1985;125(1):1–15. doi: 10.1086/284325. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
