Microglia-derived exosomes modulate myelin regeneration via miR-615-5p/MYRF axis
- PMID: 38246987
- PMCID: PMC10801965
- DOI: 10.1186/s12974-024-03019-5
Microglia-derived exosomes modulate myelin regeneration via miR-615-5p/MYRF axis
Abstract
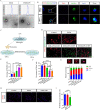
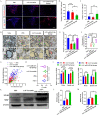
Demyelination and failure of remyelination in the central nervous system (CNS) characterize a number of neurological disorders. Spontaneous remyelination in demyelinating diseases is limited, as oligodendrocyte precursor cells (OPCs), which are often present in demyelinated lesions in abundance, mostly fail to differentiate into oligodendrocytes, the myelinating cells in the CNS. In addition to OPCs, the lesions are assembled numbers of activated resident microglia/infiltrated macrophages; however, the mechanisms and potential role of interactions between the microglia/macrophages and OPCs are poorly understood. Here, we generated a transcriptional profile of exosomes from activated microglia, and found that miR-615-5p was elevated. miR-615-5p bound to 3'UTR of myelin regulator factor (MYRF), a crucial myelination transcription factor expressed in oligodendrocyte lineage cells. Mechanistically, exosomes from activated microglia transferred miR-615-5p to OPCs, which directly bound to MYRF and inhibited OPC maturation. Furthermore, an effect of AAV expressing miR-615-5p sponge in microglia was tested in experimental autoimmune encephalomyelitis (EAE) and cuprizone (CPZ)-induced demyelination model, the classical mouse models of multiple sclerosis. miR-615-5p sponge effectively alleviated disease progression and promoted remyelination. This study identifies miR-615-5p/MYRF as a new target for the therapy of demyelinating diseases.
Keywords: Demyelinating disease; Exosomes; Microglia; OPCs; miR-615-5p.
© 2024. The Author(s).
Conflict of interest statement
We confirm this manuscript is original and has not been published before. We confirm that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted. The authors declare that they have no conflicts of interest to this work.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

