The dopamine receptor D1 inhibitor, SKF83566, suppresses GBM stemness and invasion through the DRD1-c-Myc-UHRF1 interactions
- PMID: 38246990
- PMCID: PMC10801958
- DOI: 10.1186/s13046-024-02947-7
The dopamine receptor D1 inhibitor, SKF83566, suppresses GBM stemness and invasion through the DRD1-c-Myc-UHRF1 interactions
Abstract
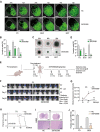
Background: Extensive local invasion of glioblastoma (GBM) cells within the central nervous system (CNS) is one factor that severely limits current treatments. The aim of this study was to uncover genes involved in the invasion process, which could also serve as therapeutic targets. For the isolation of invasive GBM cells from non-invasive cells, we used a three-dimensional organotypic co-culture system where glioma stem cell (GSC) spheres were confronted with brain organoids (BOs). Using ultra-low input RNA sequencing (ui-RNA Seq), an invasive gene signature was obtained that was exploited in a therapeutic context.
Methods: GFP-labeled tumor cells were sorted from invasive and non-invasive regions within co-cultures. Ui-RNA sequencing analysis was performed to find a gene cluster up-regulated in the invasive compartment. This gene cluster was further analyzed using the Connectivity MAP (CMap) database. This led to the identification of SKF83566, an antagonist of the D1 dopamine receptor (DRD1), as a candidate therapeutic molecule. Knockdown and overexpression experiments were performed to find molecular pathways responsible for the therapeutic effects of SKF83566. Finally, the effects of SKF83566 were validated in orthotopic xenograft models in vivo.
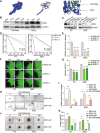
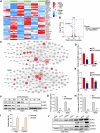
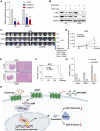
Results: Ui-RNA seq analysis of three GSC cell models (P3, BG5 and BG7) yielded a set of 27 differentially expressed genes between invasive and non-invasive cells. Using CMap analysis, SKF83566 was identified as a selective inhibitor targeting both DRD1 and DRD5. In vitro studies demonstrated that SKF83566 inhibited tumor cell proliferation, GSC sphere formation, and invasion. RNA sequencing analysis of SKF83566-treated P3, BG5, BG7, and control cell populations yielded a total of 32 differentially expressed genes, that were predicted to be regulated by c-Myc. Of these, the UHRF1 gene emerged as the most downregulated gene following treatment, and ChIP experiments revealed that c-Myc binds to its promoter region. Finally, SKF83566, or stable DRD1 knockdown, inhibited the growth of orthotopic GSC (BG5) derived xenografts in nude mice.
Conclusions: DRD1 contributes to GBM invasion and progression by regulating c-Myc entry into the nucleus that affects the transcription of the UHRF1 gene. SKF83566 inhibits the transmembrane protein DRD1, and as such represents a candidate small therapeutic molecule for GBMs.
Keywords: DRD1; Glioma stem cells; Invasion; SKF83566; UHRF1; c-Myc.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

