Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides
- PMID: 38247637
- PMCID: PMC10812719
- DOI: 10.3390/antibiotics13010078
Antibacterial and Antiviral Properties of Chenopodin-Derived Synthetic Peptides
Abstract
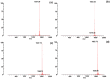
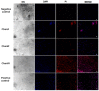
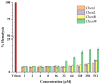
Antimicrobial peptides have been developed based on plant-derived molecular scaffolds for the treatment of infectious diseases. Chenopodin is an abundant seed storage protein in quinoa, an Andean plant with high nutritional and therapeutic properties. Here, we used computer- and physicochemical-based strategies and designed four peptides derived from the primary structure of Chenopodin. Two peptides reproduce natural fragments of 14 amino acids from Chenopodin, named Chen1 and Chen2, and two engineered peptides of the same length were designed based on the Chen1 sequence. The two amino acids of Chen1 containing amide side chains were replaced by arginine (ChenR) or tryptophan (ChenW) to generate engineered cationic and hydrophobic peptides. The evaluation of these 14-mer peptides on Staphylococcus aureus and Escherichia coli showed that Chen1 does not have antibacterial activity up to 512 µM against these strains, while other peptides exhibited antibacterial effects at lower concentrations. The chemical substitutions of glutamine and asparagine by amino acids with cationic or aromatic side chains significantly favoured their antibacterial effects. These peptides did not show significant hemolytic activity. The fluorescence microscopy analysis highlighted the membranolytic nature of Chenopodin-derived peptides. Using molecular dynamic simulations, we found that a pore is formed when multiple peptides are assembled in the membrane. Whereas, some of them form secondary structures when interacting with the membrane, allowing water translocations during the simulations. Finally, Chen2 and ChenR significantly reduced SARS-CoV-2 infection. These findings demonstrate that Chenopodin is a highly useful template for the design, engineering, and manufacturing of non-toxic, antibacterial, and antiviral peptides.
Keywords: Chenopodin; antimicrobial; antiviral; membranolytic; quinoa; synthetic peptides.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- International Monetary Fund (IMF) World Economic Outlook: Recovery during a Pandemic-Health Concerns, Supply Disruptions, Price Pressures. International Monetary Fund; Washington, DC, USA: 2021.
-
- World Health Organization (WHO) WHO Coronavirus (COVID-19) Dashboard. [(accessed on 31 December 2023)]; Available online: https://data.who.int/dashboards/covid19/cases?n=c.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

