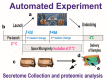
Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space
- PMID: 38247888
- PMCID: PMC10813126
- DOI: 10.3390/bioengineering11010011
Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space
Abstract
The change in gravitational force has a significant effect on biological tissues and the entire organism. As with any alteration in the environment, microgravity (µG) produces modifications in the system inducing adaptation to the new condition. In this study, we analyzed the effect of µG on neural stem cells (NSCs) following a space flight to the International Space Station (ISS). After 3 days in space, analysis of the metabolome in culture medium revealed increased glycolysis with augmented pyruvate and glycerate levels, and activated catabolism of branched-chain amino acids (BCAA) and glutamine. NSCs flown into space (SPC-NSCs) also showed increased synthesis of NADH and formation of polyamine spermidine when compared to ground controls (GC-NSCs). Overall, the space environment appears to increase energy demands in response to the µG setting.
Keywords: cell metabolism; metabolomics; microgravity; neural stem cells; secretome; spaceflight.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Space Flight Enhances Stress Pathways in Human Neural Stem Cells.Biomolecules. 2024 Jan 3;14(1):65. doi: 10.3390/biom14010065. Biomolecules. 2024. PMID: 38254665 Free PMC article.
-
Space Microgravity Alters Neural Stem Cell Division: Implications for Brain Cancer Research on Earth and in Space.Int J Mol Sci. 2022 Nov 18;23(22):14320. doi: 10.3390/ijms232214320. Int J Mol Sci. 2022. PMID: 36430810 Free PMC article.
-
Metabolomics Profile of the Secretome of Space-Flown Oligodendrocytes.Cells. 2023 Sep 11;12(18):2249. doi: 10.3390/cells12182249. Cells. 2023. PMID: 37759473 Free PMC article.
-
Spaceflight bioreactor studies of cells and tissues.Adv Space Biol Med. 2002;8:177-95. doi: 10.1016/s1569-2574(02)08019-x. Adv Space Biol Med. 2002. PMID: 12951697 Review.
-
Conducting Plant Experiments in Space and on the Moon.Methods Mol Biol. 2022;2368:165-198. doi: 10.1007/978-1-0716-1677-2_12. Methods Mol Biol. 2022. PMID: 34647256 Review.
Cited by
-
Discoveries from human stem cell research in space that are relevant to advancing cellular therapies on Earth.NPJ Microgravity. 2024 Aug 21;10(1):88. doi: 10.1038/s41526-024-00425-0. NPJ Microgravity. 2024. PMID: 39168992 Free PMC article. Review.
-
The Critical Role of YAP/BMP/ID1 Axis on Simulated Microgravity-Induced Neural Tube Defects in Human Brain Organoids.Adv Sci (Weinh). 2025 Feb;12(5):e2410188. doi: 10.1002/advs.202410188. Epub 2024 Dec 10. Adv Sci (Weinh). 2025. PMID: 39656892 Free PMC article.
References
-
- Blaber E.A., Finkelstein H., Dvorochkin N., Sato K.Y., Yousuf R., Burns B.P., Globus R.K., Almeida E.A., Oss-Ronen L., Redden R.A., et al. Microgravity Reduces the Differentiation and Regenerative Potential of Embryonic Stem Cells. Stem Cells Dev. 2015;24:2605–2621. doi: 10.1089/scd.2015.0218. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

