ResnetAge: A Resnet-Based DNA Methylation Age Prediction Method
- PMID: 38247911
- PMCID: PMC10813502
- DOI: 10.3390/bioengineering11010034
ResnetAge: A Resnet-Based DNA Methylation Age Prediction Method
Abstract
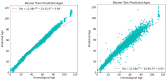
Aging is a significant contributing factor to degenerative diseases such as cancer. The extent of DNA methylation in human cells indicates the aging process and screening for age-related methylation sites can be used to construct epigenetic clocks. Thereby, it can be a new aging-detecting marker for clinical diagnosis and treatments. Predicting the biological age of human individuals is conducive to the study of physical aging problems. Although many researchers have developed epigenetic clock prediction methods based on traditional machine learning and even deep learning, higher prediction accuracy is still required to match the clinical applications. Here, we proposed an epigenetic clock prediction method based on a Resnet neuro networks model named ResnetAge. The model accepts 22,278 CpG sites as a sample input, supporting both the Illumina 27K and 450K identification frameworks. It was trained using 32 public datasets containing multiple tissues such as whole blood, saliva, and mouth. The Mean Absolute Error (MAE) of the training set is 1.29 years, and the Median Absolute Deviation (MAD) is 0.98 years. The Mean Absolute Error (MAE) of the validation set is 3.24 years, and the Median Absolute Deviation (MAD) is 2.3 years. Our method has higher accuracy in age prediction in comparison with other methylation-based age prediction methods.
Keywords: CpG sites; DNA methylation; age prediction; deep learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing.Forensic Sci Int Genet. 2017 May;28:225-236. doi: 10.1016/j.fsigen.2017.02.009. Epub 2017 Feb 28. Forensic Sci Int Genet. 2017. PMID: 28254385 Free PMC article.
-
Chronological age prediction based on DNA methylation: Massive parallel sequencing and random forest regression.Forensic Sci Int Genet. 2017 Nov;31:19-28. doi: 10.1016/j.fsigen.2017.07.015. Epub 2017 Aug 1. Forensic Sci Int Genet. 2017. PMID: 28841467
-
Human Age Prediction Based on DNA Methylation Using a Gradient Boosting Regressor.Genes (Basel). 2018 Aug 21;9(9):424. doi: 10.3390/genes9090424. Genes (Basel). 2018. PMID: 30134623 Free PMC article.
-
DNA methylation markers of age(ing) in non-model animals.Mol Ecol. 2023 Sep;32(17):4725-4741. doi: 10.1111/mec.17065. Epub 2023 Jul 3. Mol Ecol. 2023. PMID: 37401200 Review.
-
Dynamic DNA Methylation During Aging: A "Prophet" of Age-Related Outcomes.Front Genet. 2019 Feb 18;10:107. doi: 10.3389/fgene.2019.00107. eCollection 2019. Front Genet. 2019. PMID: 30833961 Free PMC article. Review.
Cited by
-
DRANetSplicer: A Splice Site Prediction Model Based on Deep Residual Attention Networks.Genes (Basel). 2024 Mar 26;15(4):404. doi: 10.3390/genes15040404. Genes (Basel). 2024. PMID: 38674339 Free PMC article.
-
Prediction of Multiple Degenerative Diseases Based on DNA Methylation in a Co-Physiology Mechanisms Perspective.Int J Mol Sci. 2024 Sep 1;25(17):9514. doi: 10.3390/ijms25179514. Int J Mol Sci. 2024. PMID: 39273460 Free PMC article.
-
The potential link between the development of Alzheimer's disease and osteoporosis.Biogerontology. 2025 Jan 20;26(1):43. doi: 10.1007/s10522-024-10181-z. Biogerontology. 2025. PMID: 39832071 Free PMC article. Review.
-
3D Modeling of Self-Expandable Valves for PPVI in Distinct RVOT Morphologies.Pediatr Cardiol. 2025 Feb 9. doi: 10.1007/s00246-025-03796-7. Online ahead of print. Pediatr Cardiol. 2025. PMID: 39923207
References
-
- De Lima C.L.P., Lapierre L.R., Singh R. A pan-tissue DNA-methylation epigenetic clock based on deep learning. NPJ Aging. 2022;8:4. doi: 10.1038/s41514-022-00085-y. - DOI
Grants and funding
- No. 62372099/National Natural Science Foundation of China
- No. 20230201090GX/Jilin Scientific and Technological Development Program
- No. 20230401092YY/Jilin Scientific and Technological Development Program
- No. YDZJ202303CGZH010/Jilin Scientific and Technological Development Program
- No. YDZJ202301ZYTS496/Jilin Scientific and Technological Development Program
LinkOut - more resources
Full Text Sources

