Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p
- PMID: 38248319
- PMCID: PMC10814901
- DOI: 10.3390/cimb46010017
Characterization of the mIF4G Domains in the RNA Surveillance Protein Upf2p
Abstract
Thirty percent of all mutations causing human disease generate mRNAs with premature termination codons (PTCs). Recognition and degradation of these PTC-containing mRNAs is carried out by the mechanism known as nonsense-mediated mRNA decay (NMD). Upf2 is a scaffold protein known to be a central component of the NMD surveillance pathway. It harbors three middle domains of eukaryotic initiation factor 4G (mIF4G-1, mIF4G-2, mIF4G-3) in its N-terminal region that are potentially important in regulating the surveillance pathway. In this study, we defined regions within the mIF4G-1 and mIF4G-2 that are required for proper function of Upf2p in NMD and translation termination in Saccharomyces cerevisiae. In addition, we narrowed down the activity of these regions to an aspartic acid (D59) in mIF4G-1 that is important for NMD activity and translation termination accuracy. Taken together, these studies suggest that inherently charged residues within mIF4G-1 of Upf2p play a role in the regulation of the NMD surveillance mechanism in S. cerevisiae.
Keywords: Saccharomyces cerevisiae; aspartic acid; cloning; codon; eukaryotic initiation factor-4G; molecular; nonsense; nonsense-mediated mRNA decay.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
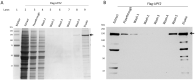









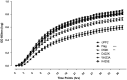
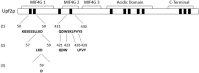
References
Grants and funding
LinkOut - more resources
Full Text Sources

