Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications
- PMID: 38249056
- PMCID: PMC10816406
- DOI: 10.3390/insects15010050
Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications
Abstract
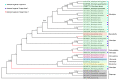
Monolepta is one of the diverse genera in the subfamily Galerucinae, including 708 species and 6 sub-species worldwide. To explore the information on the mitogenome characteristics and phylogeny of the section "Monoleptites", especially the genus Monolepta, we obtained the newly completed mitochondrial genomes (mitogenomes) of four Monolepta species using high-throughput sequencing technology. The lengths of these four new mitochondrial genomes are 16,672 bp, 16,965 bp, 16,012 bp, and 15,866 bp in size, respectively. All four mitochondrial genomes include 22 transfer RNA genes (tRNAs), 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), and one control region, which is consistent with other Coleoptera. The results of the nonsynonymous with synonymous substitution rates showed that ND6 had the highest evolution rate, while COI displayed the lowest evolution rate. The substitution saturation of three datasets (13 PCGs_codon1, 13 PCGs_codon2, 13 PCGs_codon3) showed that there was no saturation across all datasets. Phylogenetic analyses based on three datasets (ND1, 15 genes of mitogenomes, and 13 PCGs_AA) were carried out using maximum likelihood (ML) and Bayesian inference (BI) methods. The results showed that mitogenomes had a greater capacity to resolve the main clades than the ND1 gene at the suprageneric and species levels. The section "Monoleptites" was proven to be a monophyletic group, while Monolepta was a non-monophyletic group. Based on ND1 data, the newly sequenced species whose antennal segment 2 was shorter than 3 were split into several clades, while, based on the mitogenomic dataset, the four newly sequenced species had close relationships with Paleosepharia. The species whose antennal segment 2 was as long as 3 were split into two clades, which indicated that the characteristic of "antennal segment 2 as long as 3" of the true "Monolepta" evolved multiple times in several subgroups. Therefore, to explore the relationships among the true Monolepta, the most important thing is to perform a thorough revision of Monolepta and related genera in the future.
Keywords: Coleoptera; Monolepta; high-throughput sequencing; mitochondrial genome; phylogeny.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Comparative Mitogenomics in the Genus Chlorophorus (Coleoptera: Cerambycidae) and Its Phylogenetic Implications.Insects. 2024 Dec 27;16(1):8. doi: 10.3390/insects16010008. Insects. 2024. PMID: 39859589 Free PMC article.
-
The complete mitochondrial genome of Monolepta hieroglyphica (Motschulsky) (Coleoptera: Chrysomelidae).Mitochondrial DNA B Resour. 2021 Jul 15;6(8):2363-2365. doi: 10.1080/23802359.2021.1951138. eCollection 2021. Mitochondrial DNA B Resour. 2021. PMID: 34345697 Free PMC article.
-
The complete mitochondrial genome of Monolepta hieroglyphica (Motschulsky) (Coleoptera: Chrysomelidae).Mitochondrial DNA B Resour. 2021 Jun 21;6(7):2019-2021. doi: 10.1080/23802359.2021.1926363. Mitochondrial DNA B Resour. 2021. PMID: 34212083 Free PMC article.
-
Mitochondrial genome of Monolepta hieroglyphica (Coleoptera: Chrysomeloidea: Chrysomelidae) and phylogenetic analysis.Mitochondrial DNA B Resour. 2021 Apr 26;6(4):1541-1543. doi: 10.1080/23802359.2021.1914522. Mitochondrial DNA B Resour. 2021. PMID: 33969213 Free PMC article.
-
Complete mitochondrial genomes of Thyreophagus entomophagus and Acarus siro (Sarcoptiformes: Astigmatina) provide insight into mitogenome features, evolution, and phylogeny among Acaroidea mites.Exp Appl Acarol. 2022 Sep;88(1):57-74. doi: 10.1007/s10493-022-00745-4. Epub 2022 Oct 18. Exp Appl Acarol. 2022. PMID: 36255591 Review.
Cited by
-
Comparative Mitogenomics in the Genus Chlorophorus (Coleoptera: Cerambycidae) and Its Phylogenetic Implications.Insects. 2024 Dec 27;16(1):8. doi: 10.3390/insects16010008. Insects. 2024. PMID: 39859589 Free PMC article.
-
Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae).Genes (Basel). 2025 Apr 28;16(5):509. doi: 10.3390/genes16050509. Genes (Basel). 2025. PMID: 40428331 Free PMC article.
References
-
- Chevrolat L.A.A. Monolepta. In: revue, corrigée et augmentée, livraison, Dejean P.F.M.A., editor. Catalogues Des Coléopterès De La Collection De M. LeComte Dejean. Troisième éd. Volume 5. Méquignon-Marvis Père et Fils; Paris, France: 1837. pp. 385–503.
-
- Yang X.K. Genus Monolepta Chevrolat, 1837. In: Yang X.K., Ge S.Q., Nie R.E., Ruan Y.Y., Li W.Z., editors. Chinese Leaf Beetles. Science Press; Beijing, China: 2015. pp. 261–274.
-
- Zhang C., Yuan Z.H., Wang Z.Y., He K.L., Bai S.X. Population dynamics of Monolepta hieroglyphica (Motschulsky) in cornfields. J. Appl. Entomol. 2014;51:668–675.
-
- Chapuis F. Famille des Phytophages. In: Lacordaire T., Chapuis F., editors. Histoire Naturelle Des Insects. Volume 11. Genera des Coleopteres; Paris, France: 1875. pp. 1–420.
Grants and funding
LinkOut - more resources
Full Text Sources

