Rhizosphere-associated soil microbiome variability in Verticillium wilt-affected Cotinus coggygria
- PMID: 38249458
- PMCID: PMC10797040
- DOI: 10.3389/fmicb.2023.1279096
Rhizosphere-associated soil microbiome variability in Verticillium wilt-affected Cotinus coggygria
Abstract
Introduction: Verticillium wilt is the most devastating soil-borne disease affecting Cotinus coggygria in the progress of urban landscape construction in China.
Methods: To assess the variability of the rhizosphere-associated soil microbiome in response to Verticillium wilt occurrence, we investigated the microbial diversity, taxonomic composition, biomarker species, and co-occurrence network of the rhizosphere-associated soil in Verticillium wilt-affected C. coggygria using Illumina sequencing.
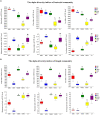
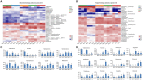
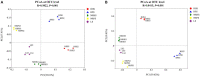
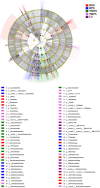
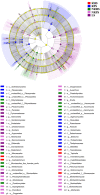
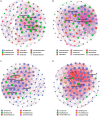
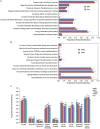
Results: The alpha diversity indices of the rhizosphere bacteria in Verticillium wilt-affected plants showed no significant variability compared with those in healthy plants, except for a moderate increase in the Shannon and Invsimpson indices, while the fungal alpha diversity indices were significantly decreased. The abundance of certain dominant or crucial microbial taxa, such as Arthrobacter, Bacillus, Streptomyces, and Trichoderma, displayed significant variations among different soil samples. The bacterial and fungal community structures exhibited distinct variability, as evidenced by the Bray-Curtis dissimilarity matrices. Co-occurrence networks unveiled intricate interactions within the microbial community of Verticillium wilt-affected C. coggygria, with greater edge numbers and higher network density. The phenomenon was more evident in the fungal community, showing increased positive interaction, which may be associated with the aggravation of Verticillium wilt with the aid of Fusarium. The proportions of bacteria involved in membrane transport and second metabolite biosynthesis functions were significantly enriched in the diseased rhizosphere soil samples.
Discussion: These findings suggested that healthy C. coggygria harbored an obviously higher abundance of beneficial microbial consortia, such as Bacillus, while Verticillium wilt-affected plants may recruit antagonistic members such as Streptomyces in response to Verticillium dahliae infection. This study provides a theoretical basis for understanding the soil micro-ecological mechanism of Verticillium wilt occurrence, which may be helpful in the prevention and control of the disease in C. coggygria from the microbiome perspective.
Keywords: Cotinus coggygria; Illumina sequencing; Verticillium wilt; plant health; rhizosphere; soil microbiome.
Copyright © 2024 Zhao, Cheng, Jiang, Qiao and Qin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Alexandra-Gabriela C., Victorita T., Cristian E. P., Elena M. M., Florenta E. H., Tatiana V. D., et al. (2023). Phenological and environmental factors' impact on secondary metabolites in medicinal plant Cotinus coggygria Scop. Plan. Theory 12:1762. doi: 10.3390/plants12091762, PMID: - DOI - PMC - PubMed
-
- Banerjee S., Kirkby C. A., Schmutter D., Bissett A., Kirkegaard J. A., Richardson A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017 - DOI
LinkOut - more resources
Full Text Sources

