Citrate metabolism in lactic acid bacteria: is there a beneficial effect for Oenococcus oeni in wine?
- PMID: 38249489
- PMCID: PMC10798043
- DOI: 10.3389/fmicb.2023.1283220
Citrate metabolism in lactic acid bacteria: is there a beneficial effect for Oenococcus oeni in wine?
Abstract
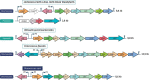
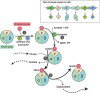
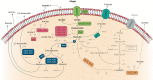
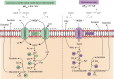
Lactic acid bacteria (LAB) are Gram positive bacteria frequently used in the food industry for fermentation, mainly transformation of carbohydrates into lactic acid. In addition, these bacteria also have the capacity to metabolize citrate, an organic acid commonly found in food products. Its fermentation leads to the production of 4-carbon compounds such as diacetyl, resulting in a buttery flavor desired in dairy products. Citrate metabolism is known to have several beneficial effects on LAB physiology. Nevertheless, a controversial effect of citrate has been described on the acid tolerance of the wine bacterium Oenococcus oeni. This observation raises questions about the effect of citrate on the capacity of O. oeni to conduct malolactic fermentation in highly acidic wines. This review aims to summarize the current understanding of citrate metabolism in LAB, with a focus on the wine bacterium O. oeni. Metabolism with the related enzymes is detailed, as are the involved genes organized in cit loci. The known systems of cit locus expression regulation are also described. Finally, the beneficial effects of citrate catabolism on LAB physiology are reported and the negative impact observed in O. oeni is discussed.
Keywords: Oenococcus oeni; citrate; citrate locus; lactic acid bacteria; metabolic engineering; proton motive force.
Copyright © 2024 Eicher, Coulon, Favier, Alexandre, Reguant and Grandvalet.
Conflict of interest statement
The authors JC and MF were employed by Biolaffort. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Alexandre H., Grandvalet C., Guilloux-Benatier M., Remize F., Tourdot-Maréchal R. (2008). Les bactéries lactiques en œnologie. Paris, France: Lavoisier.
-
- Bandell M., Lhotte M. E., Marty-Teysset C., Veyrat A., Prévost H., Dartois V., et al. (1998). Mechanism of the citrate transporters in carbohydrate and citrate cometabolism in Lactococcus and Leuconostoc species. Appl. Environ. Microbiol. 64, 1594–1600. doi: 10.1128/AEM.64.5.1594-1600.1998, PMID: - DOI - PMC - PubMed
-
- Bartowsky E. J. (2005). Oenococcus oeni and malolactic fermentation – moving into the molecular arena. Aust. J. Grape Wine Res. 11, 174–187. doi: 10.1111/j.1755-0238.2005.tb00286.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

