Comparison of differentially expressed genes in longissimus dorsi muscle of Diannan small ears, Wujin and landrace pigs using RNA-seq
- PMID: 38249550
- PMCID: PMC10796741
- DOI: 10.3389/fvets.2023.1296208
Comparison of differentially expressed genes in longissimus dorsi muscle of Diannan small ears, Wujin and landrace pigs using RNA-seq
Abstract
Introduction: Pig growth is an important economic trait that involves the co-regulation of multiple genes and related signaling pathways. High-throughput sequencing has become a powerful technology for establishing the transcriptome profiles and can be used to screen genome-wide differentially expressed genes (DEGs). In order to elucidate the molecular mechanism underlying muscle growth, this study adopted RNA sequencing (RNA-seq) to identify and compare DEGs at the genetic level in the longissimus dorsi muscle (LDM) between two indigenous Chinese pig breeds (Diannan small ears [DSE] pig and Wujin pig [WJ]) and one introduced pig breed (Landrace pig [LP]).
Methods: Animals under study were from two Chinese indigenous pig breeds (DSE pig, n = 3; WJ pig, n = 3) and one introduced pig breed (LP, n = 3) were used for RNA sequencing (RNA-seq) to identify and compare the expression levels of DEGs in the LDM. Then, functional annotation, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Protein-Protein Interaction (PPI) network analysis were performed on these DEGs. Then, functional annotation, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Protein-Protein Interaction (PPI) network analysis were performed on these DEGs.
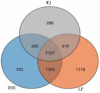
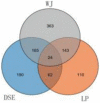
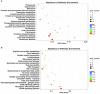
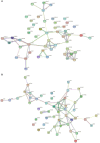
Results: The results revealed that for the DSE, WJ, and LP libraries, more than 66, 65, and 71 million clean reads were generated by transcriptome sequencing, respectively. A total of 11,213 genes were identified in the LDM tissue of these pig breeds, of which 7,127 were co-expressed in the muscle tissue of the three samples. In total, 441 and 339 DEGs were identified between DSE vs. WJ and LP vs. DSE in the study, with 254, 193 up-regulated genes and 187, 193 down-regulated genes in DSE compared to WJ and LP. GO analysis and KEGG signaling pathway analysis showed that DEGs are significantly related to contractile fiber, sarcolemma, and dystrophin-associated glycoprotein complex, myofibril, sarcolemma, and myosin II complex, Glycolysis/Gluconeogenesis, Propanoate metabolism, and Pyruvate metabolism, etc. In combination with functional annotation of DEGs, key genes such as ENO3 and JUN were identified by PPI network analysis.
Discussion: In conclusion, the present study revealed key genes including DES, FLNC, PSMD1, PSMD6, PSME4, PSMB4, RPL11, RPL13A, ROS23, RPS29, MYH1, MYL9, MYL12B, TPM1, TPM4, ENO3, PGK1, PKM2, GPI, and the unannotated new gene ENSSSCG00000020769 and related signaling pathways that influence the difference in muscle growth and could provide a theoretical basis for improving pig muscle growth traits in the future.
Keywords: Diannan small ears pig; Wujin pig; differentially expressed genes; landrace pig; longissimus dorsi muscle; transcriptome sequencing (RNA-seq).
Copyright © 2024 Li, Hao, Zhu, Yi, Cheng, Xie and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
MicroRNA analysis of porcine muscle tissue involved in phosphoinositol metabolism.Front Vet Sci. 2025 Jul 23;12:1482031. doi: 10.3389/fvets.2025.1482031. eCollection 2025. Front Vet Sci. 2025. PMID: 40777826 Free PMC article.
-
Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue.PLoS One. 2015 Oct 27;10(10):e0141138. doi: 10.1371/journal.pone.0141138. eCollection 2015. PLoS One. 2015. PMID: 26505482 Free PMC article.
-
Transcriptome analysis of differential gene expression in the longissimus dorsi muscle from Debao and landrace pigs based on RNA-sequencing.Biosci Rep. 2019 Dec 20;39(12):BSR20192144. doi: 10.1042/BSR20192144. Biosci Rep. 2019. PMID: 31755521 Free PMC article.
-
Min pig skeletal muscle response to cold stress.PLoS One. 2022 Sep 26;17(9):e0274184. doi: 10.1371/journal.pone.0274184. eCollection 2022. PLoS One. 2022. PMID: 36155652 Free PMC article. Review.
-
Comparison of muscle structure and transcriptome analysis of eyed-side muscle and blind-side muscle in Cynoglossussemilaevis (Osteichthyes, Cynoglossidae).Zookeys. 2025 Mar 6;1230:213-229. doi: 10.3897/zookeys.1230.139837. eCollection 2025. Zookeys. 2025. PMID: 40093692 Free PMC article. Review.
Cited by
-
Transcriptome Profiling Reveals Genetic Basis of Muscle Development and Meat Quality Traits in Chinese Congjiang Xiang and Landrace Pigs.Metabolites. 2025 Jun 22;15(7):426. doi: 10.3390/metabo15070426. Metabolites. 2025. PMID: 40710526 Free PMC article.
-
Analysis of genomic copy number variations through whole-genome scan in Yunling cattle.Front Vet Sci. 2024 Jul 22;11:1413504. doi: 10.3389/fvets.2024.1413504. eCollection 2024. Front Vet Sci. 2024. PMID: 39104544 Free PMC article.
-
Comparative Analysis of Short-Chain Fatty Acids and the Immune Barrier in Cecum of Dahe Pigs and Dahe Black Pigs.Animals (Basel). 2025 Mar 23;15(7):920. doi: 10.3390/ani15070920. Animals (Basel). 2025. PMID: 40218314 Free PMC article.
-
Modulating the RPS27A/PSMD12/NF-κB pathway to control immune response in mouse brain ischemia-reperfusion injury.Mol Med. 2024 Jul 22;30(1):106. doi: 10.1186/s10020-024-00870-3. Mol Med. 2024. PMID: 39039432 Free PMC article.
-
MicroRNA analysis of porcine muscle tissue involved in phosphoinositol metabolism.Front Vet Sci. 2025 Jul 23;12:1482031. doi: 10.3389/fvets.2025.1482031. eCollection 2025. Front Vet Sci. 2025. PMID: 40777826 Free PMC article.
References
-
- Wang H, Wang X, Yan D, Sun H, Chen Q, Li M, et al. . Genome-wide association study identifying genetic variants associated with carcass backfat thickness, lean percentage and fat percentage in a four-way crossbred pig population using SLAF-seq technology. BMC Genomics. (2022) 23:594. doi: 10.1186/s12864-022-08827-8, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

