Mechanistic analysis of Th2-type inflammatory factors in asthma
- PMID: 38249931
- PMCID: PMC10797403
- DOI: 10.21037/jtd-23-1628
Mechanistic analysis of Th2-type inflammatory factors in asthma
Abstract
Background: The main pathological features of asthma are widespread chronic inflammation of the airways and restricted ventilation due to airway remodeling, which involves changes in a range of regulatory pathways. While the role of T helper type 2 (Th2)-related inflammatory factors in this process is known, the detailed understanding of how genes affect protein functions during airway remodeling is still lacking. This study aims to fill this knowledge gap by integrating gene expression data and protein function analysis, providing new scientific insights for a deeper understanding of the mechanisms of airway remodeling and for further development of asthma treatment strategies.
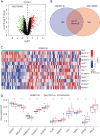
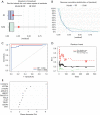
Methods: In this study, the mechanism of Th2-related inflammatory factors in tracheal remodeling was studied through differentially expressed gene (DEG) screening, enrichment analysis, protein-protein interaction (PPI) network construction, machine learning, and the construction of a line graph model.
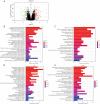
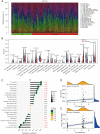
Results: Our study revealed that S100A14, KRT6A, S100A2, ABCA13, UBE2C, RASSF10, PSCA, PLAT, and TIMP1 may be the key genes for airway remodeling; epithelial-mesenchymal transition (EMT)-related genes GEM, TPM4, SLC6A8, and SNTB1 may be involved in airway remodeling due to asthma; IL6 may affect the occurrence of airway remodeling by binding to UBE2C protein or by regulating GEM genes, respectively; IL6 and IL9 may affect the occurrence of airway remodeling by regulating the downstream Toll-like receptor (TLR) signaling pathway and thus IL6 and IL9 may influence the occurrence of tracheal remodeling by regulating downstream TLR signaling pathways.
Conclusions: This study further mined the asthma gene microarray database through bioinformatics analysis and identified key genes and important pathways affecting airway remodeling in asthma patients, providing new ideas to uncover the mechanism of airway remodeling due to asthma and then seek new therapeutic targets.
Keywords: T helper type 2-associated inflammatory factors (Th2-associated inflammatory factors); airway inflammation; airway remodeling; asthma.
2023 Journal of Thoracic Disease. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1628/coif). The authors have no conflicts of interest to declare.
Figures




Similar articles
-
Targeted inhibition of β-catenin alleviates airway inflammation and remodeling in asthma via modulating the profibrotic and anti-inflammatory actions of transforming growth factor-β1.Ther Adv Respir Dis. 2021 Jan-Dec;15:1753466620981858. doi: 10.1177/1753466620981858. Ther Adv Respir Dis. 2021. PMID: 33530899 Free PMC article.
-
TMT-based quantitative proteomics revealed protective efficacy of Icariside II against airway inflammation and remodeling via inhibiting LAMP2, CTSD and CTSS expression in OVA-induced chronic asthma mice.Phytomedicine. 2023 Sep;118:154941. doi: 10.1016/j.phymed.2023.154941. Epub 2023 Jun 25. Phytomedicine. 2023. PMID: 37451150
-
TLR4 antagonist suppresses airway remodeling in asthma by inhibiting the T-helper 2 response.Exp Ther Med. 2017 Oct;14(4):2911-2916. doi: 10.3892/etm.2017.4898. Epub 2017 Aug 7. Exp Ther Med. 2017. PMID: 28966674 Free PMC article.
-
[The relation between morphologic and functional airway changes in bronchial asthma].Verh K Acad Geneeskd Belg. 2003;65(4):247-65; discussion 265-9. Verh K Acad Geneeskd Belg. 2003. PMID: 14534940 Review. Dutch.
-
Pathology of asthma.Front Microbiol. 2013 Sep 10;4:263. doi: 10.3389/fmicb.2013.00263. Front Microbiol. 2013. PMID: 24032029 Free PMC article. Review.
Cited by
-
Therapeutic potency and possible mechanism of Wuhu decoction underlying asthmatic progression via Th1/Th2 imbalance.J Thorac Dis. 2025 Jan 24;17(1):265-277. doi: 10.21037/jtd-24-804. Epub 2025 Jan 22. J Thorac Dis. 2025. PMID: 39975754 Free PMC article.
-
Tuo-Min-Ding-Chuan Decoction Alleviates Asthma via Spatial Regulation of Gut Microbiota and Treg Cell Promotion.Pharmaceuticals (Basel). 2025 Apr 28;18(5):646. doi: 10.3390/ph18050646. Pharmaceuticals (Basel). 2025. PMID: 40430465 Free PMC article.
-
Current Challenges in Pediatric Asthma.Children (Basel). 2024 May 24;11(6):632. doi: 10.3390/children11060632. Children (Basel). 2024. PMID: 38929213 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
