The genome sequence of the Willow Beauty, Peribatodes rhomboidaria (Denis & Schiffermüller, 1775)
- PMID: 38249957
- PMCID: PMC10799229
- DOI: 10.12688/wellcomeopenres.19479.1
The genome sequence of the Willow Beauty, Peribatodes rhomboidaria (Denis & Schiffermüller, 1775)
Abstract
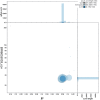
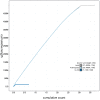
We present a genome assembly from an individual male Peribatodes rhomboidaria (the Willow Beauty; Arthropoda; Insecta; Lepidoptera; Geometridae). The genome sequence is 499.7 megabases in span. Most of the assembly is scaffolded into 31 chromosomal pseudomolecules including the Z sex chromosome. The mitochondrial genome has also been assembled and is 15.7 kilobases in length. Gene annotation of this assembly on Ensembl identified 18,486 protein coding genes.
Keywords: Lepidoptera; Peribatodes rhomboidaria; chromosomal; genome sequence; the Willow Beauty.
Copyright: © 2023 Boyes D et al.
Conflict of interest statement
No competing interests were disclosed.
Figures



Similar articles
-
The genome sequence of the Streak, Chesias legatella (Denis & Schiffermüller, 1775).Wellcome Open Res. 2024 Sep 3;8:205. doi: 10.12688/wellcomeopenres.19298.2. eCollection 2023. Wellcome Open Res. 2024. PMID: 39247705 Free PMC article.
-
The genome sequence of the Small Phoenix, Ecliptopera silaceata (Denis & Schiffermüller, 1775).Wellcome Open Res. 2023 May 10;8:209. doi: 10.12688/wellcomeopenres.19207.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39502585 Free PMC article.
-
The genome sequence of the Ashy Button, Acleris sparsana (Denis & Schiffermüller, 1775).Wellcome Open Res. 2023 Jun 12;8:241. doi: 10.12688/wellcomeopenres.19533.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38434745 Free PMC article.
-
The genome sequence of the White-point, Mythimna albipuncta (Denis & Schiffermüller, 1775).Wellcome Open Res. 2024 Feb 19;9:62. doi: 10.12688/wellcomeopenres.20682.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 38911902 Free PMC article.
-
The genome sequence of the Water Veneer, Acentria ephemerella (Denis & Schiffermüller, 1775).Wellcome Open Res. 2024 Mar 8;9:134. doi: 10.12688/wellcomeopenres.21099.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 38779149 Free PMC article.
References
-
- Busato E, Tedeschi R, Alma A: Biology and larval morphology of Peribatodes rhomboidaria (Denis & Schiffermüller) (Lepidoptera: Geometridae) on grapevine. Redia. 2000;83:11–21.
Grants and funding
LinkOut - more resources
Full Text Sources

