Identification of diagnostic molecules and potential traditional Chinese medicine components for Alzheimer's disease by single cell RNA sequencing combined with a systematic framework for network pharmacology
- PMID: 38249960
- PMCID: PMC10799563
- DOI: 10.3389/fmed.2023.1335512
Identification of diagnostic molecules and potential traditional Chinese medicine components for Alzheimer's disease by single cell RNA sequencing combined with a systematic framework for network pharmacology
Abstract
Background: Single-cell RNA sequencing (scRNA-Seq) provides new perspectives and ideas to investigate the interactions between different cell types and organisms. By integrating scRNA-seq with new computational frameworks or specific technologies, better Alzheimer's disease (AD) treatments may be developed.
Methods: The single-cell sequencing dataset GSE158234 was obtained from the GEO database. Preprocessing, quality control, dimensionality-reducing clustering, and annotation to identify cell types were performed on it. RNA-seq profiling dataset GSE238013 was used to determine the components of specific cell subpopulations in diverse samples. A set of genes included in the OMIM, Genecards, CTD, and DisGeNET databases were selected as highly plausible AD-related genes. Then, ROC curves were created to predict the diagnostic value using the significantly expressed genes in the KO group as hub genes. The genes mentioned above were mapped to the Coremine Medical database to forecast prospective therapeutic Chinese medicines, and a "Chinese medicine-ingredient-target" network was constructed to screen for potential therapeutic targets. The last step was to undertake Mendelian randomization research to determine the causal link between the critical gene IL1B and AD in the genome-wide association study.
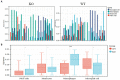
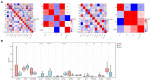
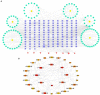
Results: Using the scRNA-seq dataset, five unique cell clusters were discovered. These clusters were further subdivided into four distinct cell types using marker genes. The KO group showed a more substantial differential subgroup of macrophages than the WT group. By using the available datasets and PPI network analysis, 54 common genes were discovered. Four clusters were identified using the MCODE approach, and correlation analysis showed that seven genes in those four clusters had a significantly negative correlation with macrophages. Six genes in four sets had a significantly positive correlation. Five genes had different levels of expression in the WT and KO groups. The String database was used to identify the regulatory relationships between the four genes (IL10, CX3CR1, IL1B, and IL6) that were finally selected as AD hub genes. Screening identified potential traditional Chinese medicine to intervene in the transformation process of AD, including Radix Salviae, ginseng, Ganoderma, licorice, Coptidis Rhizoma, and Scutellariae Radix, in addition to promising therapeutic targets, such as PTGS1, PTGS2, and RXRA. Finally, it was shown that IL1B directly correlated with immune cell infiltration in AD. In inverse variance weighting, we found that IL1B was associated with a higher risk of AD, with an OR of 1.003 (95% CI = 1.001-1.006, p = 0.038).
Conclusion: Our research combined network pharmacology and the scRNA-seq computational framework to uncover pertinent hub genes and prospective traditional Chinese medicine potential therapeutic targets for AD. These discoveries may aid in understanding the molecular processes behind AD genes and the development of novel medications to treat the condition.
Keywords: Alzheimer’s disease; Mendelian randomization; network pharmacology; single-cell transcriptome sequencing; traditional Chinese medicine.
Copyright © 2024 Wang, Zhang, Liu, Ning, Hu, Qin, Cui, Yang, Lv and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials

