Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides
- PMID: 38250930
- PMCID: PMC10818675
- DOI: 10.3390/vetsci11010024
Identification of the Linear Fc-Binding Site on the Bovine IgG1 Fc Receptor (boFcγRIII) Using Synthetic Peptides
Abstract
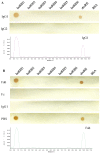
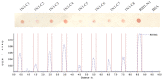
The bovine IgG1 Fc receptor (boFcγRIII) is a homologue to human FcγRIII (CD16) that binds bovine IgGI with medium-low affinity. In order to identify the Fc-binding site on the bovine IgG1 Fc receptor (boFcγRIII), peptides derived from the second extracellular domain (EC2) of boFcγRIII were synthesized and conjugated with the carrier protein. With a Dot-blot assay, the ability of the peptides to bind bovine IgG1 was determined, and the IgG1-binding peptide was also identified via truncation and mutation. The minimal peptide AQRVVN corresponding to the sequence 98-103 of boFcγRIII bound bovine IgG1 in Dot-blot, suggesting that it represents a linear ligand-binding site located in the putative A-B loop of the boFcγRIII EC2 domain. Mutation analysis of the peptide showed that the residues of Ala98, Gln99, Val101, Val102 and Asn103 within the Fc-binding site are critical for IgG1 binding on boFcγRIII. The functional peptide identified in this paper is of great value to the IgG-Fc interaction study and FcR-targeting drug development.
Keywords: BoFcγRIII; Fc-binding site; bovine IgG1; synthetic peptides.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Identification of the linear Fc-binding epitope on the bovine IgG1 Fc receptor of (boFcγRI) using synthetic peptides.J Immunol Methods. 2025 Jan;536:113799. doi: 10.1016/j.jim.2024.113799. Epub 2024 Dec 31. J Immunol Methods. 2025. PMID: 39746504
-
Identification of the linear epitope for Fc-binding on the bovine IgG2 Fc receptor (boFcgamma2R) using synthetic peptides.FEBS Lett. 2006 Feb 20;580(5):1383-90. doi: 10.1016/j.febslet.2006.01.060. Epub 2006 Jan 26. FEBS Lett. 2006. PMID: 16457820
-
Identification of a linear epitope for Fc-binding in the mouse FcγRIII.Peptides. 2010 Sep;31(9):1684-8. doi: 10.1016/j.peptides.2010.05.022. Epub 2010 Jun 8. Peptides. 2010. PMID: 20566342
-
Bovine FcgammaRIII with a single extracellular domain.Res Vet Sci. 2000 Apr;68(2):115-8. doi: 10.1053/rvsc.1999.0343. Res Vet Sci. 2000. PMID: 10756127
-
Recognition of IgG by Fcgamma receptor. The role of Fc glycosylation and the binding of peptide inhibitors.J Biol Chem. 2001 May 11;276(19):16478-83. doi: 10.1074/jbc.M100351200. Epub 2001 Jan 31. J Biol Chem. 2001. PMID: 11297533
References
-
- Bournazos S. Fc Mediated Activity of Antibodies. Volume 423. Springer; Cham, Switzerland: 2019. IgG Fc receptors: Evolutionary considerations; pp. 1–11. Current Topics in Microbiology and Immunology. - PubMed
-
- Zhu H.L., Zhao X.W., Wang X.Z., Qi Y.X., Huang D.W., Cheng G.L., Zhao H.L., Yang Y.X. Changes in expression of antimicrobial peptides and Fc receptors in the small intestines of neonatal calves during the passive immunity period. J. Dairy Sci. 2020;103:9515–9524. doi: 10.3168/jds.2019-18113. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

