Voltage-Gated Sodium Channel Inhibition by µ-Conotoxins
- PMID: 38251271
- PMCID: PMC10819908
- DOI: 10.3390/toxins16010055
Voltage-Gated Sodium Channel Inhibition by µ-Conotoxins
Abstract
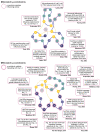
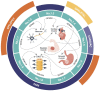
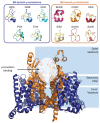
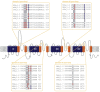
µ-Conotoxins are small, potent pore-blocker inhibitors of voltage-gated sodium (NaV) channels, which have been identified as pharmacological probes and putative leads for analgesic development. A limiting factor in their therapeutic development has been their promiscuity for different NaV channel subtypes, which can lead to undesirable side-effects. This review will focus on four areas of µ-conotoxin research: (1) mapping the interactions of µ-conotoxins with different NaV channel subtypes, (2) µ-conotoxin structure-activity relationship studies, (3) observed species selectivity of µ-conotoxins and (4) the effects of µ-conotoxin disulfide connectivity on activity. Our aim is to provide a clear overview of the current status of µ-conotoxin research.
Keywords: disulfide-rich peptide; peptide; structure–activity relationships; subtype selectivity; venom peptide; voltage-gated sodium channels; µ-conotoxin.
Conflict of interest statement
C.I.S. is an employee of Genentech Inc. and a shareholder of Roche. K.L.M. and I.V. declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

