Future Prospects, Approaches, and the Government's Role in the Development of a Hepatitis C Virus Vaccine
- PMID: 38251345
- PMCID: PMC10820710
- DOI: 10.3390/pathogens13010038
Future Prospects, Approaches, and the Government's Role in the Development of a Hepatitis C Virus Vaccine
Abstract
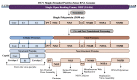
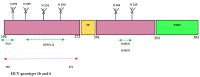
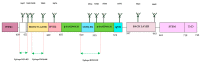
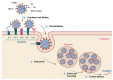
Developing a safe and effective vaccine against the hepatitis C virus (HCV) remains a top priority for global health. Despite recent advances in antiviral therapies, the high cost and limited accessibility of these treatments impede their widespread application, particularly in resource-limited settings. Therefore, the development of the HCV vaccine remains a necessity. This review article analyzes the current technologies, future prospects, strategies, HCV genomic targets, and the governmental role in HCV vaccine development. We discuss the current epidemiological landscape of HCV infection and the potential of HCV structural and non-structural protein antigens as vaccine targets. In addition, the involvement of government agencies and policymakers in supporting and facilitating the development of HCV vaccines is emphasized. We explore how vaccine development regulatory channels and frameworks affect research goals, funding, and public health policy. The significance of international and public-private partnerships in accelerating the development of an HCV vaccine is examined. Finally, the future directions for developing an HCV vaccine are discussed. In conclusion, the review highlights the urgent need for a preventive vaccine to fight the global HCV disease and the significance of collaborative efforts between scientists, politicians, and public health organizations to reach this important public health goal.
Keywords: COVID-19; SARS-CoV-2; government agencies; hepatitis C virus; prevalence; vaccine.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
References
-
- El-Zanaty F., Way A. Egypt Demographic and Health Survey 2008. Ministry of Health; Cairo, Egypt: 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

