Concurrent Tissue and Circulating Tumor DNA Molecular Profiling to Detect Guideline-Based Targeted Mutations in a Multicancer Cohort
- PMID: 38252441
- PMCID: PMC10804266
- DOI: 10.1001/jamanetworkopen.2023.51700
Concurrent Tissue and Circulating Tumor DNA Molecular Profiling to Detect Guideline-Based Targeted Mutations in a Multicancer Cohort
Abstract
Importance: Tissue-based next-generation sequencing (NGS) of solid tumors is the criterion standard for identifying somatic mutations that can be treated with National Comprehensive Cancer Network guideline-recommended targeted therapies. Sequencing of circulating tumor DNA (ctDNA) can also identify tumor-derived mutations, and there is increasing clinical evidence supporting ctDNA testing as a diagnostic tool. The clinical value of concurrent tissue and ctDNA profiling has not been formally assessed in a large, multicancer cohort from heterogeneous clinical settings.
Objective: To evaluate whether patients concurrently tested with both tissue and ctDNA NGS testing have a higher rate of detection of guideline-based targeted mutations compared with tissue testing alone.
Design, setting, and participants: This cohort study comprised 3209 patients who underwent sequencing between May 2020, and December 2022, within the deidentified, Tempus multimodal database, consisting of linked molecular and clinical data. Included patients had stage IV disease (non-small cell lung cancer, breast cancer, prostate cancer, or colorectal cancer) with sufficient tissue and blood sample quantities for analysis.
Exposures: Received results from tissue and plasma ctDNA genomic profiling, with biopsies and blood draws occurring within 30 days of one another.
Main outcomes and measures: Detection rates of guideline-based variants found uniquely by ctDNA and tissue profiling.
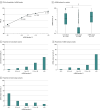
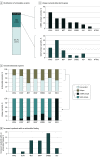
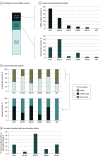
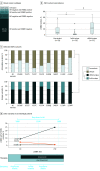
Results: The cohort of 3209 patients (median age at diagnosis of stage IV disease, 65.3 years [2.5%-97.5% range, 43.3-83.3 years]) who underwent concurrent tissue and ctDNA testing included 1693 women (52.8%). Overall, 1448 patients (45.1%) had a guideline-based variant detected. Of these patients, 9.3% (135 of 1448) had variants uniquely detected by ctDNA profiling, and 24.2% (351 of 1448) had variants uniquely detected by solid-tissue testing. Although largely concordant with one another, differences in the identification of actionable variants by either assay varied according to cancer type, gene, variant, and ctDNA burden. Of 352 patients with breast cancer, 20.2% (71 of 352) with actionable variants had unique findings in ctDNA profiling results. Most of these unique, actionable variants (55.0% [55 of 100]) were found in ESR1, resulting in a 24.7% increase (23 of 93) in the identification of patients harboring an ESR1 mutation relative to tissue testing alone.
Conclusions and relevance: This study suggests that unique actionable biomarkers are detected by both concurrent tissue and ctDNA testing, with higher ctDNA identification among patients with breast cancer. Integration of concurrent NGS testing into the routine management of advanced solid cancers may expand the delivery of molecularly guided therapy and improve patient outcomes.
Conflict of interest statement
Figures

Comment in
-
Concurrent Circulating Tumor DNA and Tissue Genotyping-Ready for Prime Time?JAMA Netw Open. 2024 Jan 2;7(1):e2351679. doi: 10.1001/jamanetworkopen.2023.51679. JAMA Netw Open. 2024. PMID: 38252442 No abstract available.
References
-
- National Comprehensive Cancer Network . Breast cancer (version 3.2023). Accessed March 22, 2023. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
-
- National Comprehensive Cancer Network . Non-small cell lung cancer (version 2.2023). Accessed March 22, 2023. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
-
- National Comprehensive Cancer Network . Prostate cancer (version 1.2023). Accessed March 22, 2023. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
-
- National Comprehensive Cancer Network . Colon cancer (version 1.2023). Accessed April 13, 2023. https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
-
- National Comprehensive Cancer Network . Rectal cancer (version 1.2023). Accessed April 13, 2023. https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

