PreS1BP mediates inhibition of Hepatitis B virus replication by promoting HBx protein degradation
- PMID: 38253259
- PMCID: PMC10846407
- DOI: 10.1016/j.virusres.2024.199326
PreS1BP mediates inhibition of Hepatitis B virus replication by promoting HBx protein degradation
Abstract
Background: PreS1-binding protein (PreS1BP), recognized as a nucleolar protein and tumor suppressor, influences the replication of various viruses, including vesicular stomatitis virus (VSV) and herpes simplex virus type 1 (HSV-1). Its role in hepatitis B virus (HBV) replication and the underlying mechanisms, however, remain elusive.
Methods: We investigated PreS1BP expression levels in an HBV-replicating cell and animal model and analyzed the impact of its overexpression on viral replication metrics. HBV DNA, covalently closed circular DNA (cccDNA), hepatitis B surface antigen (HBsAg), hepatitis B core antigen (HBcAg), and HBV RNA levels were assessed in HBV-expressing stable cell lines under varying PreS1BP conditions. Furthermore, co-immunoprecipitation and ubiquitination assays were used to detect PreS1BP- hepatitis B virus X protein (HBx) interactions and HBx stability modulated by PreS1BP.
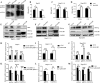
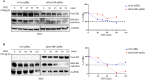
Results: Our study revealed a marked decrease in PreS1BP expression in the presence of active HBV replication. Functional assays showed that PreS1BP overexpression significantly inhibited HBV replication and transcription, evidenced by the reduction in HBV DNA, cccDNA, HBsAg, HBcAg, and HBV RNA levels. At the molecular level, PreS1BP facilitated the degradation of HBx in a dose-dependent fashion, whereas siRNA-mediated knockdown of PreS1BP led to an increase in HBx levels. Subsequent investigations uncovered that PreS1BP accelerated HBx protein degradation via K63-linked ubiquitination in a ubiquitin-proteasome system-dependent manner. Co-immunoprecipitation assays further established that PreS1BP enhances the recruitment of the proteasome 20S subunit alpha 3 (PSMA3) for interaction with HBx, thereby fostering its degradation.
Conclusions: These findings unveil a previously unidentified mechanism wherein PreS1BP mediates HBx protein degradation through the ubiquitin-proteasome system, consequentially inhibiting HBV replication. This insight positions PreS1BP as a promising therapeutic target for future HBV interventions. Further studies are warranted to explore the clinical applicability of modulating PreS1BP in HBV therapy.
Keywords: HBx; Hepatitis B virus; PreS1BP; Ubiquitination.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner.J Virol. 2019 Mar 5;93(6):e01840-18. doi: 10.1128/JVI.01840-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567987 Free PMC article.
-
Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx.J Hepatol. 2021 Mar;74(3):522-534. doi: 10.1016/j.jhep.2020.09.019. Epub 2020 Sep 25. J Hepatol. 2021. PMID: 32987030
-
Lost Small Envelope Protein Expression from Naturally Occurring PreS1 Deletion Mutants of Hepatitis B Virus Is Often Accompanied by Increased HBx and Core Protein Expression as Well as Genome Replication.J Virol. 2021 Jun 24;95(14):e0066021. doi: 10.1128/JVI.00660-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33910956 Free PMC article.
-
SMC5/6-Mediated Transcriptional Regulation of Hepatitis B Virus and Its Therapeutic Potential.Viruses. 2024 Oct 25;16(11):1667. doi: 10.3390/v16111667. Viruses. 2024. PMID: 39599784 Free PMC article. Review.
-
Decoding the multifaceted interventions between human sirtuin 2 and dynamic hepatitis B viral proteins to confirm their roles in HBV replication.Front Cell Infect Microbiol. 2024 Jan 4;13:1234903. doi: 10.3389/fcimb.2023.1234903. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38239506 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

