Short-duration selective decontamination of the digestive tract infection control does not contribute to increased antimicrobial resistance burden in a pilot cluster randomised trial (the ARCTIC Study)
- PMID: 38253478
- PMCID: PMC11103307
- DOI: 10.1136/gutjnl-2023-330851
Short-duration selective decontamination of the digestive tract infection control does not contribute to increased antimicrobial resistance burden in a pilot cluster randomised trial (the ARCTIC Study)
Abstract
Objective: Selective decontamination of the digestive tract (SDD) is a well-studied but hotly contested medical intervention of enhanced infection control. Here, we aim to characterise the changes to the microbiome and antimicrobial resistance (AMR) gene profiles in critically ill children treated with SDD-enhanced infection control compared with conventional infection control.
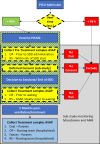
Design: We conducted shotgun metagenomic microbiome and resistome analysis on serial oropharyngeal and faecal samples collected from critically ill, mechanically ventilated patients in a pilot multicentre cluster randomised trial of SDD. The microbiome and AMR profiles were compared for longitudinal and intergroup changes. Of consented patients, faecal microbiome baseline samples were obtained in 89 critically ill children. Additionally, samples collected during and after critical illness were collected in 17 children treated with SDD-enhanced infection control and 19 children who received standard care.
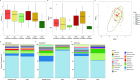
Results: SDD affected the alpha and beta diversity of critically ill children to a greater degree than standard care. At cessation of treatment, the microbiome of SDD patients was dominated by Actinomycetota, specifically Bifidobacterium, at the end of mechanical ventilation. Altered gut microbiota was evident in a subset of SDD-treated children who returned late longitudinal samples compared with children receiving standard care. Clinically relevant AMR gene burden was unaffected by the administration of SDD-enhanced infection control compared with standard care. SDD did not affect the composition of the oral microbiome compared with standard treatment.
Conclusion: Short interventions of SDD caused a shift in the microbiome but not of the AMR gene pool in critically ill children at the end mechanical ventilation, compared with standard antimicrobial therapy.
Keywords: ANTIBIOTIC THERAPY; DRUG RESISTANCE.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: IRLK was funded by Action for Medical Research. NP holds grants from Action for Medical Research, the NIHR and Addenbrookes Charitable Trust (ACT). SB is funded by the Wellcome Trust.
Figures



Comment in
-
Selective decontamination of the digestive tract in critically ill children: fighting fire with fire or burning down the house?Gut. 2024 May 10;73(6):883-884. doi: 10.1136/gutjnl-2024-331955. Gut. 2024. PMID: 38519126 No abstract available.
References
-
- Northway T, Langley JM, Skippen P. Health care–associated infection in the pediatric intensive care unit. Pediatr Crit Care 2011:1349–63.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
