Self-renewing human naïve pluripotent stem cells dedifferentiate in 3D culture and form blastoids spontaneously
- PMID: 38253551
- PMCID: PMC10803796
- DOI: 10.1038/s41467-024-44969-x
Self-renewing human naïve pluripotent stem cells dedifferentiate in 3D culture and form blastoids spontaneously
Abstract
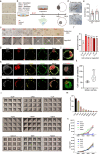
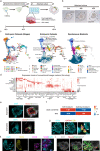
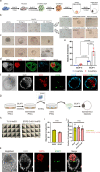
Human naïve pluripotent stem cells (hnPSCs) can generate integrated models of blastocysts termed blastoids upon switch to inductive medium. However, the underlying mechanisms remain obscure. Here we report that self-renewing hnPSCs spontaneously and efficiently give rise to blastoids upon three dimensional (3D) suspension culture. The spontaneous blastoids mimic early stage human blastocysts in terms of structure, size, and transcriptome characteristics and are capable of progressing to post-implantation stages. This property is conferred by the glycogen synthase kinase-3 (GSK3) signalling inhibitor IM-12 present in 5iLAF self-renewing medium. IM-12 upregulates oxidative phosphorylation-associated genes that underly the capacity of hnPSCs to generate blastoids spontaneously. Starting from day one of self-organization, hnPSCs at the boundary of all 3D aggregates dedifferentiate into E5 embryo-like intermediates. Intermediates co-express SOX2/OCT4 and GATA6 and by day 3 specify trophoblast fate, which coincides with cavity and blastoid formation. In summary, spontaneous blastoid formation results from 3D culture triggering dedifferentiation of hnPSCs into earlier embryo-like intermediates which are then competent to segregate blastocyst fates.
© 2024. The Author(s).
Conflict of interest statement
J.C.R.S., M.G., J.W., C.C., and K.W. submitted a patent application (202211174246.1) related to this work. Other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

