LOX-1 acts as an N6-methyladenosine-regulated receptor for Helicobacter pylori by binding to the bacterial catalase
- PMID: 38253620
- PMCID: PMC10803311
- DOI: 10.1038/s41467-024-44860-9
LOX-1 acts as an N6-methyladenosine-regulated receptor for Helicobacter pylori by binding to the bacterial catalase
Abstract
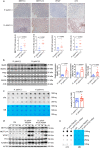
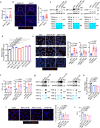
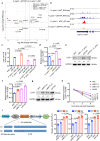
The role of N6-methyladenosine (m6A) modification of host mRNA during bacterial infection is unclear. Here, we show that Helicobacter pylori infection upregulates host m6A methylases and increases m6A levels in gastric epithelial cells. Reducing m6A methylase activity via hemizygotic deletion of methylase-encoding gene Mettl3 in mice, or via small interfering RNAs targeting m6A methylases, enhances H. pylori colonization. We identify LOX-1 mRNA as a key m6A-regulated target during H. pylori infection. m6A modification destabilizes LOX-1 mRNA and reduces LOX-1 protein levels. LOX-1 acts as a membrane receptor for H. pylori catalase and contributes to bacterial adhesion. Pharmacological inhibition of LOX-1, or genetic ablation of Lox-1, reduces H. pylori colonization. Moreover, deletion of the bacterial catalase gene decreases adhesion of H. pylori to human gastric sections. Our results indicate that m6A modification of host LOX-1 mRNA contributes to protection against H. pylori infection by downregulating LOX-1 and thus reducing H. pylori adhesion.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

