An improved pathway for autonomous bioluminescence imaging in eukaryotes
- PMID: 38253843
- PMCID: PMC10927554
- DOI: 10.1038/s41592-023-02152-y
An improved pathway for autonomous bioluminescence imaging in eukaryotes
Abstract
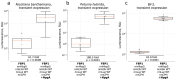
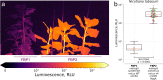
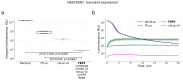
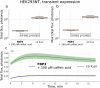
The discovery of the bioluminescence pathway in the fungus Neonothopanus nambi enabled engineering of eukaryotes with self-sustained luminescence. However, the brightness of luminescence in heterologous hosts was limited by performance of the native fungal enzymes. Here we report optimized versions of the pathway that enhance bioluminescence by one to two orders of magnitude in plant, fungal and mammalian hosts, and enable longitudinal video-rate imaging.
© 2024. The Author(s).
Conflict of interest statement
This study was partially funded by Light Bio (light.bio) and Planta (planta.bio). K.V.W. is the CEO of Light Bio. K.S.S., K.V.W. and I.V.Y. are shareholders of Light Bio. A.S.M. is the CSO of Planta. E.S.S., T.A.K., N.M.M., T.M., K.A.P., M.M.P., L.I.F., A.K.M., E.N.B., L.K.P., A.V.B. and V.V.C. are employees of Planta. A.E.B., D.A.G., V.V.B., D.I.B., A.Y.G., F.G., M.G.W., T.T.H., M.P.H. and L.P.E. declare no competing interests.
Figures








References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

