Variant of the lactase LCT gene explains association between milk intake and incident type 2 diabetes
- PMID: 38253929
- PMCID: PMC11097298
- DOI: 10.1038/s42255-023-00961-1
Variant of the lactase LCT gene explains association between milk intake and incident type 2 diabetes
Abstract
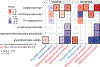
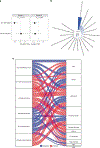
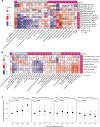
Cow's milk is frequently included in the human diet, but the relationship between milk intake and type 2 diabetes (T2D) remains controversial. Here, using data from the Hispanic Community Health Study/Study of Latinos, we show that in both sexes, higher milk intake is associated with lower risk of T2D in lactase non-persistent (LNP) individuals (determined by a variant of the lactase LCT gene, single nucleotide polymorphism rs4988235 ) but not in lactase persistent individuals. We validate this finding in the UK Biobank. Further analyses reveal that among LNP individuals, higher milk intake is associated with alterations in gut microbiota (for example, enriched Bifidobacterium and reduced Prevotella) and circulating metabolites (for example, increased indolepropionate and reduced branched-chain amino acid metabolites). Many of these metabolites are related to the identified milk-associated bacteria and partially mediate the association between milk intake and T2D in LNP individuals. Our study demonstrates a protective association between milk intake and T2D among LNP individuals and a potential involvement of gut microbiota and blood metabolites in this association.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


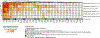







References
-
- Pereira PC Milk nutritional composition and its role in human health. Nutrition 30, 619–627 (2014). - PubMed
-
- Gijsbers L et al. Consumption of dairy foods and diabetes incidence: a dose-response meta-analysis of observational studies. Am. J. Clin. Nutr. 103, 1111–1124 (2016). - PubMed
-
- Segurel L & Bon C On the evolution of lactase persistence in humans. Annu Rev. Genomics Hum. Genet. 18, 297–319 (2017). - PubMed
-
- Storhaug CL, Fosse SK & Fadnes LT Country, regional, and global estimates for lactose malabsorption in adults: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2, 738–746 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- R00 DK122128/DK/NIDDK NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- U54 GM104940/GM/NIGMS NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- R01 HL140976/HL/NHLBI NIH HHS/United States
- R01 HL142003/HL/NHLBI NIH HHS/United States
- R01 HL136266/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- R01 DK120870/DK/NIDDK NIH HHS/United States
- R01 DK126698/DK/NIDDK NIH HHS/United States
- N01 HC065233/HL/NHLBI NIH HHS/United States
- R01 MD011389/MD/NIMHD NIH HHS/United States
- N01 HC065236/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- N01 HC065235/HL/NHLBI NIH HHS/United States
- R01 DK119268/DK/NIDDK NIH HHS/United States
- P30 DK111022/DK/NIDDK NIH HHS/United States
- N01 HC065234/HL/NHLBI NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- R01 HL141824/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- R01 HL060712/HL/NHLBI NIH HHS/United States
- UM1 HG008898/HG/NHGRI NIH HHS/United States
- R01 DK132011/DK/NIDDK NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- N01 HC065237/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

