TEM1/endosialin/CD248 promotes pathologic scarring and TGF-β activity through its receptor stability in dermal fibroblasts
- PMID: 38254097
- PMCID: PMC10804696
- DOI: 10.1186/s12929-024-01001-0
TEM1/endosialin/CD248 promotes pathologic scarring and TGF-β activity through its receptor stability in dermal fibroblasts
Abstract
Background: Pathologic scars, including keloids and hypertrophic scars, represent a common form of exaggerated cutaneous scarring that is difficult to prevent or treat effectively. Additionally, the pathobiology of pathologic scars remains poorly understood. We aim at investigating the impact of TEM1 (also known as endosialin or CD248), which is a glycosylated type I transmembrane protein, on development of pathologic scars.
Methods: To investigate the expression of TEM1, we utilized immunofluorescence staining, Western blotting, and single-cell RNA-sequencing (scRNA-seq) techniques. We conducted in vitro cell culture experiments and an in vivo stretch-induced scar mouse model to study the involvement of TEM1 in TGF-β-mediated responses in pathologic scars.
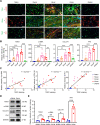
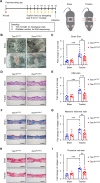
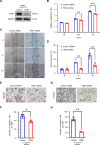
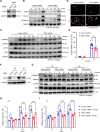
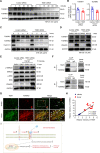
Results: The levels of the protein TEM1 are elevated in both hypertrophic scars and keloids in comparison to normal skin. A re-analysis of scRNA-seq datasets reveals that a major profibrotic subpopulation of keloid and hypertrophic scar fibroblasts greatly expresses TEM1, with expression increasing during fibroblast activation. TEM1 promotes activation, proliferation, and ECM production in human dermal fibroblasts by enhancing TGF-β1 signaling through binding with and stabilizing TGF-β receptors. Global deletion of Tem1 markedly reduces the amount of ECM synthesis and inflammation in a scar in a mouse model of stretch-induced pathologic scarring. The intralesional administration of ontuxizumab, a humanized IgG monoclonal antibody targeting TEM1, significantly decreased both the size and collagen density of keloids.
Conclusions: Our data indicate that TEM1 plays a role in pathologic scarring, with its synergistic effect on the TGF-β signaling contributing to dermal fibroblast activation. Targeting TEM1 may represent a novel therapeutic approach in reducing the morbidity of pathologic scars.
Keywords: Fibroblast activation; Hypertrophic scar; Keloid; TEM1/endosialin/CD248; TGF-β.
© 2024. The Author(s).
Conflict of interest statement
The authors state no conflict of interest.
Figures


References
-
- Altman N, Krzywinski M. Clustering. Nat Methods. 2017;14(6):545–546. doi: 10.1038/nmeth.4299. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

