Prevalence and genetic evolution of porcine reproductive and respiratory syndrome virus in commercial fattening pig farms in China
- PMID: 38254191
- PMCID: PMC10801985
- DOI: 10.1186/s40813-024-00356-y
Prevalence and genetic evolution of porcine reproductive and respiratory syndrome virus in commercial fattening pig farms in China
Abstract
Background: To investigate the prevalence and evolution of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) at commercial fattening pig farms, a total of 1397 clinical samples were collected from a single fattening cycle at seven pig farms in five provinces of China from 2020 to 2021.
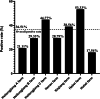
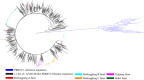
Results: The RT‒PCR results revealed that PRRSV was present on all seven farms, and the percentage of PRRSV-positive individuals was 17.54-53.33%. A total of 344 partial NSP2 gene sequences and 334 complete ORF5 gene sequences were obtained from the positive samples. The statistical results showed that PRRSV-2 was present on all seven commercial fattening farms, and PRRSV-1 was present on only one commercial fattening farm. A total of six PRRSV-2 subtypes were detected, and five of the seven farms had two or more PRRSV-2 subtypes. L1.8 (L1C) PRRSV was the dominant epidemic strain on five of the seven pig farms. Sequence analysis of L1.8 (L1C) PRRSV from different commercial fattening pig farms revealed that its consistency across farms varied substantially. The amino acid alignment results demonstrated that there were 131 aa discontinuous deletions in NSP2 between different L1.8 (L1C) PRRSV strains and that the GP5 mutation in L1.8 (L1C) PRRSV was mainly concentrated in the peptide signal region and T-cell epitopes. Selection pressure analysis of GP5 revealed that the use of the PRRSV MLV vaccine had no significant episodic diversifying effect on L1.8 (L1C) PRRSV.
Conclusion: PRRSV infection is common at commercial fattening pig farms in China, and the percentage of positive individuals is high. There are multiple PRRSV subtypes of infection at commercial fattening pig farms in China. L1.8 (L1C) is the main circulating PRRSV strain on commercial fattening pig farms. L1.8 (L1C) PRRSV detected at different commercial fattening pig farms exhibited substantial differences in consistency but similar molecular characteristics. The pressure on the GP5 of L1.8 (L1C) PRRSV may not be directly related to the use of the vaccines.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Zhang WL, Zhang HL, Xu H, Tang YD, Leng CL, Peng JM, Wang Q, An TQ, Cai XH, Fan JH, Tian ZJ. Two novel recombinant porcine reproductive and respiratory syndrome viruses belong to sublineage 35 originating from sublineage 32. Transbound Emerg Dis. 2019;66(6):2592–2600. doi: 10.1111/tbed.13320. - DOI - PubMed
Grants and funding
- GZC20233062/National funded postdoctoral researcher program
- SKLVBF202208/State Key Laboratory of Veterinary Biotechnology Foundation
- 2022YFF0711004/National Key Research and Development Program
- 32172890/National Natural Science Foundation of China
- 32002315/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous

