Antimicrobial resistance crisis: could artificial intelligence be the solution?
- PMID: 38254241
- PMCID: PMC10804841
- DOI: 10.1186/s40779-024-00510-1
Antimicrobial resistance crisis: could artificial intelligence be the solution?
Abstract
Antimicrobial resistance is a global public health threat, and the World Health Organization (WHO) has announced a priority list of the most threatening pathogens against which novel antibiotics need to be developed. The discovery and introduction of novel antibiotics are time-consuming and expensive. According to WHO's report of antibacterial agents in clinical development, only 18 novel antibiotics have been approved since 2014. Therefore, novel antibiotics are critically needed. Artificial intelligence (AI) has been rapidly applied to drug development since its recent technical breakthrough and has dramatically improved the efficiency of the discovery of novel antibiotics. Here, we first summarized recently marketed novel antibiotics, and antibiotic candidates in clinical development. In addition, we systematically reviewed the involvement of AI in antibacterial drug development and utilization, including small molecules, antimicrobial peptides, phage therapy, essential oils, as well as resistance mechanism prediction, and antibiotic stewardship.
Keywords: Antibiotic; Antibiotic stewardship; Antimicrobial peptide; Artificial intelligence (AI); Clinical development; Machine learning (ML); Phage therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no interests of conflicts.
Figures


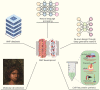
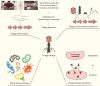

References
-
- World Health Organization. A financial model for an impact investment fund for the development of antibacterial treatments and diagnostics: a user guide. 2020. Available from: https://www.who.int/publications/i/item/a-financial-model-for-an-impact-.... Accessed 1 Jun 2020.
-
- Centers for Disease Control and Prevention (U.S.). Antibiotic resistance threats in the United States, 2013. Atlanta, GA: US Department of Health and Human Services. 2013. Available from: https://stacks.cdc.gov/view/cdc/20705.
-
- CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: US Department of Health and Human Services. 2019. Available from: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re....
-
- O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial resistance. 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

