Dynamic Evolution of Repetitive Elements and Chromatin States in Apis mellifera Subspecies
- PMID: 38254978
- PMCID: PMC10815273
- DOI: 10.3390/genes15010089
Dynamic Evolution of Repetitive Elements and Chromatin States in Apis mellifera Subspecies
Abstract
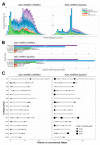
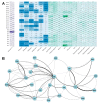
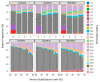
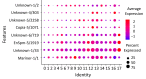
In this study, we elucidate the contribution of repetitive DNA sequences to the establishment of social structures in honeybees (Apis mellifera). Despite recent advancements in understanding the molecular mechanisms underlying the formation of honeybee castes, primarily associated with Notch signaling, the comprehensive identification of specific genomic cis-regulatory sequences remains elusive. Our objective is to characterize the repetitive landscape within the genomes of two honeybee subspecies, namely A. m. mellifera and A. m. ligustica. An observed recent burst of repeats in A. m. mellifera highlights a notable distinction between the two subspecies. After that, we transitioned to identifying differentially expressed DNA elements that may function as cis-regulatory elements. Nevertheless, the expression of these sequences showed minimal disparity in the transcriptome during caste differentiation, a pivotal process in honeybee eusocial organization. Despite this, chromatin segmentation, facilitated by ATAC-seq, ChIP-seq, and RNA-seq data, revealed a distinct chromatin state associated with repeats. Lastly, an analysis of sequence divergence among elements indicates successive changes in repeat states, correlating with their respective time of origin. Collectively, these findings propose a potential role of repeats in acquiring novel regulatory functions.
Keywords: Hymenoptera; TE); chromatin landscape; honeybee Apis mellifera; noncoding RNA (ncRNA); repetitive DNA; transposable elements (transposons.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Christmann S. Do We Realize the Full Impact of Pollinator Loss on Other Ecosystem Services and the Challenges for Any Restoration in Terrestrial Areas? Restor. Ecol. 2019;27:720–725. doi: 10.1111/rec.12950. - DOI
-
- Dangles O., Casas J. Ecosystem Services Provided by Insects for Achieving Sustainable Development Goals. Ecosyst. Serv. 2019;35:109–115. doi: 10.1016/j.ecoser.2018.12.002. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

