MicroAnnot: A Dedicated Workflow for Accurate Microsporidian Genome Annotation
- PMID: 38255958
- PMCID: PMC10815200
- DOI: 10.3390/ijms25020880
MicroAnnot: A Dedicated Workflow for Accurate Microsporidian Genome Annotation
Abstract
With nearly 1700 species, Microsporidia represent a group of obligate intracellular eukaryotes with veterinary, economic and medical impacts. To help understand the biological functions of these microorganisms, complete genome sequencing is routinely used. Nevertheless, the proper prediction of their gene catalogue is challenging due to their taxon-specific evolutionary features. As innovative genome annotation strategies are needed to obtain a representative snapshot of the overall lifestyle of these parasites, the MicroAnnot tool, a dedicated workflow for microsporidian sequence annotation using data from curated databases of accurately annotated microsporidian genes, has been developed. Furthermore, specific modules have been implemented to perform small gene (<300 bp) and transposable element identification. Finally, functional annotation was performed using the signature-based InterProScan software. MicroAnnot's accuracy has been verified by the re-annotation of four microsporidian genomes for which structural annotation had previously been validated. With its comparative approach and transcriptional signal identification method, MicroAnnot provides an accurate prediction of translation initiation sites, an efficient identification of transposable elements, as well as high specificity and sensitivity for microsporidian genes, including those under 300 bp.
Keywords: dedicated workflow; high quality annotation; microsporidia; structural annotation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
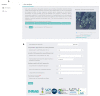
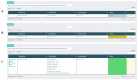
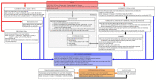
References
-
- Ruan Y., Xu X., He Q., Li L., Guo J., Bao J., Pan G., Li T., Zhou Z. The Largest Meta-Analysis on the Global Prevalence of Microsporidia in Mammals, Avian and Water Provides Insights into the Epidemic Features of These Ubiquitous Pathogens. Parasites Vectors. 2021;14:186. doi: 10.1186/s13071-021-04700-x. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

