Identification and Expression Analysis of the Nucleotidyl Transferase Protein (NTP) Family in Soybean (Glycine max) under Various Abiotic Stresses
- PMID: 38256188
- PMCID: PMC10816777
- DOI: 10.3390/ijms25021115
Identification and Expression Analysis of the Nucleotidyl Transferase Protein (NTP) Family in Soybean (Glycine max) under Various Abiotic Stresses
Abstract
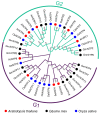
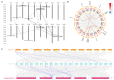
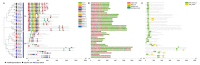
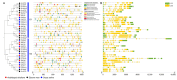
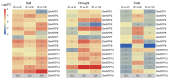
Nucleotidyl transferases (NTPs) are common transferases in eukaryotes and play a crucial role in nucleotide modifications at the 3' end of RNA. In plants, NTPs can regulate RNA stability by influencing 3' end modifications, which in turn affect plant growth, development, stress responses, and disease resistance. Although the functions of NTP family members have been extensively studied in Arabidopsis, rice, and maize, there is limited knowledge about NTP genes in soybeans. In this study, we identified 16 members of the NTP family in soybeans, including two subfamilies (G1 and G2) with distinct secondary structures, conserved motifs, and domain distributions at the protein level. Evolutionary analysis of genes in the NTP family across multiple species and gene collinearity analysis revealed a relatively conserved evolutionary pattern. Analysis of the tertiary structure of the proteins showed that NTPs have three conserved aspartic acids that bind together to form a possible active site. Tissue-specific expression analysis indicated that some NTP genes exhibit tissue-specific expression, likely due to their specific functions. Stress expression analysis showed significant differences in the expression levels of NTP genes under high salt, drought, and cold stress. Additionally, RNA-seq analysis of soybean plants subjected to salt and drought stress further confirmed the association of soybean NTP genes with abiotic stress responses. Subcellular localization experiments revealed that GmNTP2 and GmNTP14, which likely have similar functions to HESO1 and URT1, are located in the nucleus. These research findings provide a foundation for further investigations into the functions of NTP family genes in soybeans.
Keywords: Glycine max; RNA-seq; abiotic stress; gene expression; nucleotidyl transferase protein (NTP); subcellular localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Functions and mechanisms of RNA tailing by nucleotidyl transferase proteins in plants.Front Plant Sci. 2024 Oct 15;15:1452347. doi: 10.3389/fpls.2024.1452347. eCollection 2024. Front Plant Sci. 2024. PMID: 39474218 Free PMC article. Review.
-
Identification and expression profiling of Oryza sativa nucleotidyl transferase protein (NTP) genes under various stress conditions.Gene. 2017 Sep 10;628:93-102. doi: 10.1016/j.gene.2017.06.038. Epub 2017 Jul 1. Gene. 2017. PMID: 28676446
-
Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress.BMC Genomics. 2014 Nov 3;15:950. doi: 10.1186/1471-2164-15-950. BMC Genomics. 2014. PMID: 25362847 Free PMC article.
-
Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis.PLoS Genet. 2015 Apr 30;11(4):e1005119. doi: 10.1371/journal.pgen.1005119. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25928405 Free PMC article.
-
Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression.Plant J. 2015 Feb;81(3):505-18. doi: 10.1111/tpj.12746. Plant J. 2015. PMID: 25495120
Cited by
-
Integrated transcriptomic and metabolomic analyses provide insights into defense against Colletotrichum fructicola in octoploid strawberries.BMC Plant Biol. 2025 Feb 13;25(1):190. doi: 10.1186/s12870-025-06057-0. BMC Plant Biol. 2025. PMID: 39948459 Free PMC article.
-
Functions and mechanisms of RNA tailing by nucleotidyl transferase proteins in plants.Front Plant Sci. 2024 Oct 15;15:1452347. doi: 10.3389/fpls.2024.1452347. eCollection 2024. Front Plant Sci. 2024. PMID: 39474218 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

