The Role of Protein Methyltransferases in Immunity
- PMID: 38257273
- PMCID: PMC10819338
- DOI: 10.3390/molecules29020360
The Role of Protein Methyltransferases in Immunity
Abstract
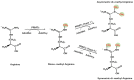
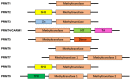
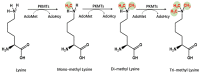
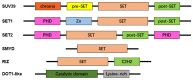
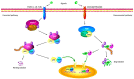
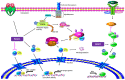
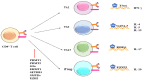
The immune system protects our body from bacteria, viruses, and toxins and removes malignant cells. Activation of immune cells requires the onset of a network of important signaling proteins. Methylation of these proteins affects their structure and biological function. Under stimulation, T cells, B cells, and other immune cells undergo activation, development, proliferation, differentiation, and manufacture of cytokines and antibodies. Methyltransferases alter the above processes and lead to diverse outcomes depending on the degree and type of methylation. In the previous two decades, methyltransferases have been reported to mediate a great variety of immune stages. Elucidating the roles of methylation in immunity not only contributes to understanding the immune mechanism but is helpful in the development of new immunotherapeutic strategies. Hence, we review herein the studies on methylation in immunity, aiming to provide ideas for new approaches.
Keywords: arginine methylation; inflammation; lysine methylation; protein methylation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation.Nat Commun. 2017 Oct 12;8(1):891. doi: 10.1038/s41467-017-01009-1. Nat Commun. 2017. PMID: 29026071 Free PMC article.
-
Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity.Biochemistry. 2004 Aug 24;43(33):10834-43. doi: 10.1021/bi049626i. Biochemistry. 2004. PMID: 15311944
-
[Progress in the study of histone methyltransferases].Yi Chuan. 2007 Sep;29(9):1035-41. doi: 10.1360/yc-007-1035. Yi Chuan. 2007. PMID: 17855250 Review. Chinese.
-
Everything is E(Z): linking histone methylation to B cell development.Nat Immunol. 2003 Feb;4(2):101-3. doi: 10.1038/ni0203-101. Nat Immunol. 2003. PMID: 12555091 No abstract available.
-
Structure and function of histone methyltransferases.Crit Rev Eukaryot Gene Expr. 2004;14(3):147-69. doi: 10.1615/critreveukaryotgeneexpr.v14.i3.10. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15248813 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

