The Repurposing of Cellular Proteins during Enterovirus A71 Infection
- PMID: 38257775
- PMCID: PMC10821071
- DOI: 10.3390/v16010075
The Repurposing of Cellular Proteins during Enterovirus A71 Infection
Abstract
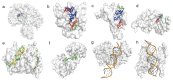
Viruses pose a great threat to people's lives. Enterovirus A71 (EV-A71) infects children and infants all over the world with no FDA-approved treatment to date. Understanding the basic mechanisms of viral processes aids in selecting more efficient drug targets and designing more effective antivirals to thwart this virus. The 5'-untranslated region (5'-UTR) of the viral RNA genome is composed of a cloverleaf structure and an internal ribosome entry site (IRES). Cellular proteins that bind to the cloverleaf structure regulate viral RNA synthesis, while those that bind to the IRES also known as IRES trans-acting factors (ITAFs) regulate viral translation. In this review, we survey the cellular proteins currently known to bind the 5'-UTR and influence viral gene expression with emphasis on comparing proteins' functions and localizations pre- and post-(EV-A71) infection. A comprehensive understanding of how the host cell's machinery is hijacked and reprogrammed by the virus to facilitate its replication is crucial for developing effective antivirals.
Keywords: 5′-UTR; IRES; ITAF; enterovirus A71 (EV-A71).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


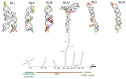

References
-
- Enterovirus 71. [(accessed on 8 November 2023)]. Available online: https://www.who.int/teams/health-product-policy-and-standards/standards-....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

