Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany
- PMID: 38257802
- PMCID: PMC10818819
- DOI: 10.3390/v16010102
Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany
Abstract
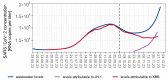
The importance of COVID-19 surveillance from wastewater continues to grow since case-based surveillance in the general population has been scaled back world-wide. In Berlin, Germany, quantitative and genomic wastewater monitoring for SARS-CoV-2 is performed in three wastewater treatment plants (WWTP) covering 84% of the population since December 2021. The SARS-CoV-2 Omicron sublineage JN.1 (B.2.86.1.1), was first identified from wastewater on 22 October 2023 and rapidly became the dominant sublineage. This change was accompanied by a parallel and still ongoing increase in the notification-based 7-day-hospitalization incidence of COVID-19 and COVID-19 ICU utilization, indicating increasing COVID-19 activity in the (hospital-prone) population and a higher strain on the healthcare system. In retrospect, unique mutations of JN.1 could be identified in wastewater as early as September 2023 but were of unknown relevance at the time. The timely detection of new sublineages in wastewater therefore depends on the availability of new sequences from GISAID and updates to Pango lineage definitions and Nextclade. We show that genomic wastewater surveillance provides timely public health evidence on a regional level, complementing the existing indicators.
Keywords: Germany; Omicron; SARS-CoV-2; genomic surveillance; wastewater sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Zhakparov D., Quirin Y., Xiao Y., Battaglia N., Holzer M., Bühler M., Kistler W., Engel D., Zumthor J.P., Caduff A., et al. Sequencing of SARS-CoV-2 RNA Fragments in Wastewater Detects the Spread of New Variants during Major Events. Microorganisms. 2023;11:2660. doi: 10.3390/microorganisms11112660. - DOI - PMC - PubMed
-
- McManus O., Christiansen L.E., Nauta M., Krogsgaard L.W., Bahrenscheer N.S., Von Kappelgaard L., Christiansen T., Hansen M., Hansen N.C., Kähler J., et al. Predicting COVID-19 Incidence Using Wastewater Surveillance Data, Denmark, October 2021–June 2022. Emerg. Infect. Dis. 2023;29:1589–1597. doi: 10.3201/eid2908.221634. - DOI - PMC - PubMed
-
- Espinosa-Gongora C., Berg C., Rehn M., Varg J.E., Dillner L., Latorre-Margalef N., Székely A.J., Andersson E., Movert E. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage through Integrated Genomic Surveillance of Wastewater and COVID-19 Cases in Sweden, Weeks 31 to 38 2023. Eurosurveillance. 2023;28:2300595. doi: 10.2807/1560-7917.ES.2023.28.46.2300595. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

