Narrowing the diagnostic gap: Genomes, episignatures, long-read sequencing, and health economic analyses in an exome-negative intellectual disability cohort
- PMID: 38258669
- PMCID: PMC11786952
- DOI: 10.1016/j.gim.2024.101076
Narrowing the diagnostic gap: Genomes, episignatures, long-read sequencing, and health economic analyses in an exome-negative intellectual disability cohort
Abstract
Purpose: Genome sequencing (GS)-specific diagnostic rates in prospective tightly ascertained exome sequencing (ES)-negative intellectual disability (ID) cohorts have not been reported extensively.
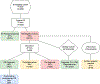
Methods: ES, GS, epigenetic signatures, and long-read sequencing diagnoses were assessed in 74 trios with at least moderate ID.
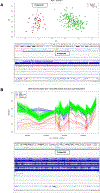
Results: The ES diagnostic yield was 42 of 74 (57%). GS diagnoses were made in 9 of 32 (28%) ES-unresolved families. Repeated ES with a contemporary pipeline on the GS-diagnosed families identified 8 of 9 single-nucleotide variations/copy-number variations undetected in older ES, confirming a GS-unique diagnostic rate of 1 in 32 (3%). Episignatures contributed diagnostic information in 9% with GS corroboration in 1 of 32 (3%) and diagnostic clues in 2 of 32 (6%). A genetic etiology for ID was detected in 51 of 74 (69%) families. Twelve candidate disease genes were identified. Contemporary ES followed by GS cost US$4976 (95% CI: $3704; $6969) per diagnosis and first-line GS at a cost of $7062 (95% CI: $6210; $8475) per diagnosis.
Conclusion: Performing GS only in ID trios would be cost equivalent to ES if GS were available at $2435, about a 60% reduction from current prices. This study demonstrates that first-line GS achieves higher diagnostic rate than contemporary ES but at a higher cost.
Keywords: Episignature; Exome negative; Genome sequencing; Health economics; Intellectual disability.
Copyright © 2024 American College of Medical Genetics and Genomics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures
References
-
- Srivastava S, Love-Nichols JA, Dies KA, Ledbetter DH, Martin CL, Chung WK, Firth HV, Frazier T, Hansen RL, Prock L, et al. (2019). Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genetics in Medicine. 10.1038/s41436-019-0554-6. - DOI - PMC - PubMed
-
- Marshall CR, Chowdhury S, Taft RJ, Lebo MS, Buchan JG, Harrison SM, Rowsey R, Klee EW, Liu P, Worthey EA, et al. (2020). Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease. NPJ Genom Med 5. 10.1038/s41525-020-00154-9. - DOI - PMC - PubMed
-
- Ewans LJ, Schofield D, Shrestha R, Zhu Y, Gayevskiy V, Ying K, Walsh C, Lee E, Kirk EP, Colley A, et al. (2018). Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genetics in Medicine 20, 1564–1574. 10.1038/gim.2018.39. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

