Farnesoid X receptor mediates macrophage-intrinsic responses to suppress colitis-induced colon cancer progression
- PMID: 38258906
- PMCID: PMC10906220
- DOI: 10.1172/jci.insight.170428
Farnesoid X receptor mediates macrophage-intrinsic responses to suppress colitis-induced colon cancer progression
Abstract
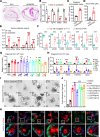
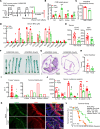
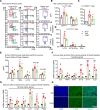
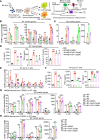
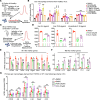
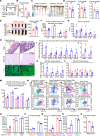
Bile acids (BAs) affect the intestinal environment by ensuring barrier integrity, maintaining microbiota balance, regulating epithelium turnover, and modulating the immune system. As a master regulator of BA homeostasis, farnesoid X receptor (FXR) is severely compromised in patients with inflammatory bowel disease (IBD) and colitis-associated colorectal cancer (CAC). At the front line, gut macrophages react to the microbiota and metabolites that breach the epithelium. We aim to study the role of the BA/FXR axis in macrophages. This study demonstrates that inflammation-induced epithelial abnormalities compromised FXR signaling and altered BAs' profile in a mouse CAC model. Further, gut macrophage-intrinsic FXR sensed aberrant BAs, leading to pro-inflammatory cytokines' secretion, which promoted intestinal stem cell proliferation. Mechanistically, activation of FXR ameliorated intestinal inflammation and inhibited colitis-associated tumor growth, by regulating gut macrophages' recruitment, polarization, and crosstalk with Th17 cells. However, deletion of FXR in bone marrow or gut macrophages escalated the intestinal inflammation. In summary, our study reveals a distinctive regulatory role of FXR in gut macrophages, suggesting its potential as a therapeutic target for addressing IBD and CAC.
Keywords: Colorectal cancer; Endocrinology; Gastroenterology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

