Anaerobic gut fungal communities in marsupial hosts
- PMID: 38259066
- PMCID: PMC10865811
- DOI: 10.1128/mbio.03370-23
Anaerobic gut fungal communities in marsupial hosts
Abstract
The anaerobic gut fungi (AGF) inhabit the alimentary tracts of herbivores. In contrast to placental mammals, information regarding the identity, diversity, and community structure of AGF in marsupials is extremely sparse. Here, we characterized AGF communities in 61 fecal samples from 10 marsupial species belonging to four families in the order Diprotodontia: Vombatidae (wombats), Phascolarctidae (koalas), Phalangeridae (possums), and Macropodidae (kangaroos, wallabies, and pademelons). An amplicon-based diversity survey using the D2 region of the large ribosomal subunit as a phylogenetic marker indicated that marsupial AGF communities were dominated by eight genera commonly encountered in placental herbivores (Neocallimastix, Caecomyces, Cyllamyces, Anaeromyces, Orpinomyces, Piromyces, Pecoramyces, and Khoyollomyces). Community structure analysis revealed a high level of stochasticity, and ordination approaches did not reveal a significant role for the animal host, gut type, dietary preferences, or lifestyle in structuring marsupial AGF communities. Marsupial foregut and hindgut communities displayed diversity and community structure patterns comparable to AGF communities typically encountered in placental foregut hosts while exhibiting a higher level of diversity and a distinct community structure compared to placental hindgut communities. Quantification of AGF load using quantitative PCR indicated a significantly smaller load in marsupial hosts compared to their placental counterparts. Isolation efforts were only successful from a single red kangaroo fecal sample and yielded a Khoyollomyces ramosus isolate closely related to strains previously isolated from placental hosts. Our results suggest that AGF communities in marsupials are in low abundance and show little signs of selection based on ecological and evolutionary factors.IMPORTANCEThe AGF are integral part of the microbiome of herbivores. They play a crucial role in breaking down plant biomass in hindgut and foregut fermenters. The majority of research has been conducted on the AGF community in placental mammalian hosts. However, it is important to note that many marsupial mammals are also herbivores and employ a hindgut or foregut fermentation strategy for breaking down plant biomass. So far, very little is known regarding the AGF diversity and community structure in marsupial mammals. To fill this knowledge gap, we conducted an amplicon-based diversity survey targeting AGF in 61 fecal samples from 10 marsupial species. We hypothesize that, given the distinct evolutionary history and alimentary tract architecture, novel and unique AGF communities would be encountered in marsupials. Our results indicate that marsupial AGF communities are highly stochastic, present in relatively low loads, and display community structure patterns comparable to AGF communities typically encountered in placental foregut hosts. Our results indicate that marsupial hosts harbor AGF communities; however, in contrast to the strong pattern of phylosymbiosis typically observed between AGF and placental herbivores, the identity and gut architecture appear to play a minor role in structuring AGF communities in marsupials.
Keywords: anaerobic fungi; community structure; marsupials.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



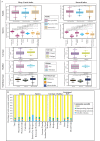



References
-
- Arman SD, Prideaux GJ. 2015. Dietary classification of extant kangaroos and their relatives (Marsupialia: Macropodoidea). Austral Ecol 40:909–922. doi:10.1111/aec.12273 - DOI
-
- Melzer A, Cristescu R, Ellis W, FitzGibbon S, Manno G. 2014. The habitat and diet of koalas (Phascolarctos cinereus) in Queensland. Aust Mammal 36:189. doi:10.1071/AM13032 - DOI
-
- Freeman MS. 2018. Marsupial diet, p 1–8. In Vonk J, Shackelford T (ed), Encyclopedia of animal cognition and behavior. Springer International Publishing, Cham.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
