Identification and characterization of methoxy- and dimethoxyhydroquinone 1,2-dioxygenase from Phanerochaete chrysosporium
- PMID: 38259078
- PMCID: PMC10880611
- DOI: 10.1128/aem.01753-23
Identification and characterization of methoxy- and dimethoxyhydroquinone 1,2-dioxygenase from Phanerochaete chrysosporium
Abstract
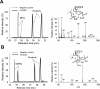
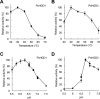
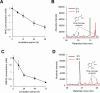
White-rot fungi, such as Phanerochaete chrysosporium, are the most efficient degraders of lignin, a major component of plant biomass. Enzymes produced by these fungi, such as lignin peroxidases and manganese peroxidases, break down lignin polymers into various aromatic compounds based on guaiacyl, syringyl, and hydroxyphenyl units. These intermediates are further degraded, and the aromatic ring is cleaved by 1,2,4-trihydroxybenzene dioxygenases. This study aimed to characterize homogentisate dioxygenase (HGD)-like proteins from P. chrysosporium that are strongly induced by the G-unit fragment of vanillin. We overexpressed two homologous recombinant HGDs, PcHGD1 and PcHGD2, in Escherichia coli. Both PcHGD1 and PcHGD2 catalyzed the ring cleavage in methoxyhydroquinone (MHQ) and dimethoxyhydroquinone (DMHQ). The two enzymes had the highest catalytic efficiency (kcat/Km) for MHQ, and therefore, we named PcHGD1 and PcHGD2 as MHQ dioxygenases 1 and 2 (PcMHQD1 and PcMHQD2), respectively, from P. chrysosporium. This is the first study to identify and characterize MHQ and DMHQ dioxygenase activities in members of the HGD superfamily. These findings highlight the unique and broad substrate spectra of PcHGDs, rendering them attractive candidates for biotechnological applications.IMPORTANCEThis study aimed to elucidate the properties of enzymes responsible for degrading lignin, a dominant natural polymer in terrestrial lignocellulosic biomass. We focused on two homogentisate dioxygenase (HGD) homologs from the white-rot fungus, P. chrysosporium, and investigated their roles in the degradation of lignin-derived aromatic compounds. In the P. chrysosporium genome database, PcMHQD1 and PcMHQD2 were annotated as HGDs that could cleave the aromatic rings of methoxyhydroquinone (MHQ) and dimethoxyhydroquinone (DMHQ) with a preference for MHQ. These findings suggest that MHQD1 and/or MHQD2 play important roles in the degradation of lignin-derived aromatic compounds by P. chrysosporium. The preference of PcMHQDs for MHQ and DMHQ not only highlights their potential for biotechnological applications but also underscores their critical role in understanding lignin degradation by a representative of white-rot fungus, P. chrysosporium.
Keywords: homogentisate dioxygenase; lignin; syringic acid; vanillic acid; white-rot fungus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sarkanen KV, Ludwig CH. 1971. Lignins: occurrence, formation, structure and reactions. J Polym Sci B Polym Phys 10:228–230.
-
- Crawford RL. 1982. Lignin biodegradation and transformation. Enzyme Microb Technol 4:285. doi:10.1016/0141-0229(82)90047-3 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

