A novel conjugative transposon carrying an autonomously amplified plasmid
- PMID: 38259081
- PMCID: PMC10865816
- DOI: 10.1128/mbio.02787-23
A novel conjugative transposon carrying an autonomously amplified plasmid
Abstract
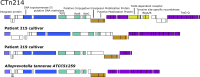
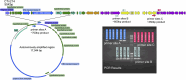
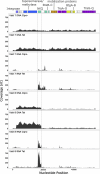
Tetracyclines serve as broad-spectrum antibiotics to treat bacterial infections. The discovery of new tetracycline resistance genes has led to new questions about the underlying mechanisms of resistance, gene transfer, and their relevance to human health. We tracked changes in the abundance of a 55-kbp conjugative transposon (CTn214) carrying tetQ, a tetracycline resistance gene, within a Bacteroides fragilis metagenome-assembled genome derived from shotgun sequencing of microbial DNA extracted from the ileal pouch of a patient with ulcerative colitis. The mapping of metagenomic reads to CTn214 revealed the multi-copy nature of a 17,044-nt region containing tetQ in samples collected during inflammation and uninflamed visits. B. fragilis cultivars isolated from the same patient during periods of inflammation harbored CTn214 integrated into the chromosome or both a circular, multi-copy, extrachromosomal region of the CTn214 containing tetQ and the corresponding integrated form. The tetracycline-dependent mechanism for the transmission of CTn214 is nearly identical to a common conjugative transposon found in the genome of B. fragilis (CTnDOT), but the autonomously amplified nature of a circular 17,044-nt region of CTn214 that codes for tetQ and the integration of the same sequence in the linear chromosome within the same cell is a novel observation. Genome and transcriptome sequencing of B. fragilis cultivars grown under different concentrations of tetracycline and ciprofloxacin indicates that tetQ in strains containing the circular form remains actively expressed regardless of treatment, while the expression of tetQ in strains containing the linear form increases only in the presence of tetracycline.IMPORTANCEThe exchange of antibiotic production and resistance genes between microorganisms can lead to the emergence of new pathogens. In this study, short-read mapping of metagenomic samples taken over time from the illeal pouch of a patient with ulcerative colitis to a Bacteroides fragilis metagenome-assembled genome revealed two distinct genomic arrangements of a novel conjugative transposon, CTn214, that encodes tetracycline resistance. The autonomous amplification of a plasmid-like circular form from CTn214 that includes tetQ potentially provides consistent ribosome protection against tetracycline. This mode of antibiotic resistance offers a novel mechanism for understanding the emergence of pathobionts like B. fragilis and their persistence for extended periods of time in patients with inflammatory bowel disease.
Keywords: Bacteroides fragilis; CTnDOT; antibiotic resistance; conjugal transposon; host–microbe interactions; microbial evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Guérillot R, Da Cunha V, Sauvage E, Bouchier C, Glaser P. 2013. Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication, and conjugation for their spreading. J Bacteriol 195:1979–1990. doi:10.1128/JB.01745-12 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
