The complete chloroplast genome sequence of the North American sclerophyllous evergreen shrub, Quercus turbinella (Fagaceae)
- PMID: 38259356
- PMCID: PMC10802804
- DOI: 10.1080/23802359.2024.2305398
The complete chloroplast genome sequence of the North American sclerophyllous evergreen shrub, Quercus turbinella (Fagaceae)
Abstract
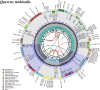
Quercus turbinella (section Quercus; Fagaceae) is an evergreen shrub characteristic in central Arizona and it concerns one of the most abundant and economically important genera of Quercus in the Northern Hemisphere. Here, we have sequenced the complete chloroplast genome to provide insight into the phylogenetic relationship of Q. turbinella. The whole genome is 161,208 bp in length with two inverted repeat regions of 25,827 bp each, which separate a large single-copy region of 90,552 bp and a small single-copy region of 19,002 bp. A total of 136 genes were annotated, including 88 protein-coding genes, eight ribosomal RNAs, and 40 transfer RNAs. The result of the maximum-likelihood phylogenetic analysis strongly suggested that Quercus turbinella had a close relationship to Quercus macrocarpa with strong bootstrap support.
Keywords: Chloroplast genome; Quercus turbinella; phylogenetic analysis.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Holmgren CA, Betancourt JL, Rylander KA.. 2011. Vegetation history along the eastern, desert escarpment of the Sierra San Pedro Mártir, Baja California, Mexico. Quat Res. 75(3):647–657. doi:10.1016/j.yqres.2011.01.008. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
