Exploring the memory: existing activity-dependent tools to tag and manipulate engram cells
- PMID: 38259503
- PMCID: PMC10800721
- DOI: 10.3389/fncel.2023.1279032
Exploring the memory: existing activity-dependent tools to tag and manipulate engram cells
Abstract
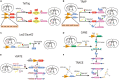
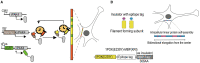
The theory of engrams, proposed several years ago, is highly crucial to understanding the progress of memory. Although it significantly contributes to identifying new treatments for cognitive disorders, it is limited by a lack of technology. Several scientists have attempted to validate this theory but failed. With the increasing availability of activity-dependent tools, several researchers have found traces of engram cells. Activity-dependent tools are based on the mechanisms underlying neuronal activity and use a combination of emerging molecular biological and genetic technology. Scientists have used these tools to tag and manipulate engram neurons and identified numerous internal connections between engram neurons and memory. In this review, we provide the background, principles, and selected examples of applications of existing activity-dependent tools. Using a combination of traditional definitions and concepts of engram cells, we discuss the applications and limitations of these tools and propose certain developmental directions to further explore the functions of engram cells.
Keywords: activity-dependent tools; engram cells; genetic strategy; memory; neuronal activity.
Copyright © 2024 Pang, Wu, Chen, Yan, Du, Yu, Yang, Wang and Lu.
Conflict of interest statement
WW was employed by Guangzhou Southern Medical Laboratory Animal Sci. and Tech. Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
An update on memory formation and retrieval: An engram-centric approach.Alzheimers Dement. 2020 Jun;16(6):926-937. doi: 10.1002/alz.12071. Epub 2020 Apr 25. Alzheimers Dement. 2020. PMID: 32333509
-
Brain-wide immunolabeling and tissue clearing applications for engram research.Neurobiol Learn Mem. 2025 Mar;218:108032. doi: 10.1016/j.nlm.2025.108032. Epub 2025 Feb 6. Neurobiol Learn Mem. 2025. PMID: 39922482 Review.
-
A Synaptic Framework for the Persistence of Memory Engrams.Front Synaptic Neurosci. 2021 Mar 24;13:661476. doi: 10.3389/fnsyn.2021.661476. eCollection 2021. Front Synaptic Neurosci. 2021. PMID: 33841124 Free PMC article. Review.
-
Memory engrams: Recalling the past and imagining the future.Science. 2020 Jan 3;367(6473):eaaw4325. doi: 10.1126/science.aaw4325. Science. 2020. PMID: 31896692 Free PMC article. Review.
-
Identification and optogenetic manipulation of memory engrams in the hippocampus.Front Behav Neurosci. 2014 Jan 17;7:226. doi: 10.3389/fnbeh.2013.00226. eCollection 2013. Front Behav Neurosci. 2014. PMID: 24478647 Free PMC article. Review.
Cited by
-
Characterization of neuronal ensembles in a model of dual reward conditioned place preference.bioRxiv [Preprint]. 2025 Jun 15:2025.06.11.659163. doi: 10.1101/2025.06.11.659163. bioRxiv. 2025. PMID: 40661519 Free PMC article. Preprint.
-
What could evolve in the evolution of memory?Philos Trans R Soc Lond B Biol Sci. 2025 Jun 26;380(1929):20240109. doi: 10.1098/rstb.2024.0109. Epub 2025 Jun 26. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40566906 Free PMC article.
-
The Blood-Cerebrospinal Fluid Barrier Dysfunction in Brain Disorders and Stroke: Why, How, What For?Neuromolecular Med. 2024 Sep 15;26(1):38. doi: 10.1007/s12017-024-08806-0. Neuromolecular Med. 2024. PMID: 39278883 Review.
References
Publication types
LinkOut - more resources
Full Text Sources

