Polyketide synthases mutation in tuberculosis transmission revealed by whole genomic sequence, China, 2011-2019
- PMID: 38259610
- PMCID: PMC10800454
- DOI: 10.3389/fgene.2023.1217255
Polyketide synthases mutation in tuberculosis transmission revealed by whole genomic sequence, China, 2011-2019
Abstract
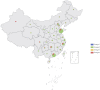
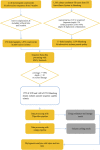
Introduction: Tuberculosis (TB) is an infectious disease caused by a bacterium called Mycobacterium tuberculosis (Mtb). Previous studies have primarily focused on the transmissibility of multidrug-resistant (MDR) or extensively drug-resistant (XDR) Mtb. However, variations in virulence across Mtb lineages may also account for differences in transmissibility. In Mtb, polyketide synthase (PKS) genes encode large multifunctional proteins which have been shown to be major mycobacterial virulence factors. Therefore, this study aimed to identify the role of PKS mutations in TB transmission and assess its risk and characteristics. Methods: Whole genome sequences (WGSs) data from 3,204 Mtb isolates was collected from 2011 to 2019 in China. Whole genome single nucleotide polymorphism (SNP) profiles were used for phylogenetic tree analysis. Putative transmission clusters (≤10 SNPs) were identified. To identify the role of PKS mutations in TB transmission, we compared SNPs in the PKS gene region between "clustered isolates" and "non-clustered isolates" in different lineages. Results: Cluster-associated mutations in ppsA, pks12, and pks13 were identified among different lineage isolates. They were statistically significant among clustered strains, indicating that they may enhance the transmissibility of Mtb. Conclusion: Overall, this study provides new insights into the function of PKS and its localization in M. tuberculosis. The study found that ppsA, pks12, and pks13 may contribute to disease progression and higher transmission of certain strains. We also discussed the prospective use of mutant ppsA, pks12, and pks13 genes as drug targets.
Keywords: Mycobacterium tuberculosis; mutation; phylogenetic analysis; polyketide synthases; transmission.
Copyright © 2024 Wang, Hu, Li, Kong, Li, Sun, Wang, Li, Zhang, Han, Zhu, An, Liu, Liu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Transmission, distribution and drug resistance-conferring mutations of extensively drug-resistant tuberculosis in the Western Cape Province, South Africa.Microb Genom. 2022 Apr;8(4):000815. doi: 10.1099/mgen.0.000815. Microb Genom. 2022. PMID: 35471145 Free PMC article.
-
High-throughput phenogenotyping of Mycobacteria tuberculosis clinical strains reveals bacterial determinants of treatment outcomes.bioRxiv [Preprint]. 2023 Apr 10:2023.04.09.536166. doi: 10.1101/2023.04.09.536166. bioRxiv. 2023. PMID: 37090677 Free PMC article. Preprint.
-
Analysis of Drug-Resistance Characteristics and Genetic Diversity of Multidrug-Resistant Tuberculosis Based on Whole-Genome Sequencing on the Hainan Island, China.Infect Drug Resist. 2023 Sep 4;16:5783-5798. doi: 10.2147/IDR.S423955. eCollection 2023. Infect Drug Resist. 2023. PMID: 37692467 Free PMC article.
-
Correlations between drug resistance of Beijing/W lineage clinical isolates of Mycobacterium tuberculosis and sublineages: a 2009-2013 prospective study in Xinjiang province, China.Med Sci Monit. 2015 May 7;21:1313-8. doi: 10.12659/MSM.892951. Med Sci Monit. 2015. PMID: 25950148 Free PMC article.
-
[Development of antituberculous drugs: current status and future prospects].Kekkaku. 2006 Dec;81(12):753-74. Kekkaku. 2006. PMID: 17240921 Review. Japanese.
Cited by
-
Genetic insights in infectious diseases: Insights from a case report and implications for personalized medicine.World J Clin Cases. 2025 May 6;13(13):101438. doi: 10.12998/wjcc.v13.i13.101438. World J Clin Cases. 2025. PMID: 40330288 Free PMC article.
References
-
- Alibaud L., Rombouts Y., Trivelli X., Burguière A., Cirillo S. L. G., Cirillo J. D., et al. (2011). A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol. Microbiol. 80 (1365-2958), 919–934. (Electronic)). 10.1111/j.1365-2958.2011.07618.x - DOI - PubMed
-
- Astarie-Dequeker C., Le Guyader L., Malaga W., Seaphanh F. K., Chalut C., Lopez A., et al. (2009). Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 5, e1000289. 1553-7374 (Electronic)). 10.1371/journal.ppat.1000289 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

