Turkey hen sperm storage tubule transcriptome response to artificial insemination and the presence of semen
- PMID: 38260096
- PMCID: PMC10801083
- DOI: 10.3389/fphys.2023.1305168
Turkey hen sperm storage tubule transcriptome response to artificial insemination and the presence of semen
Abstract
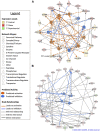
Introduction: Sperm storage within the uterovaginal junction (UVJ) of avian species occurs in specialized structures termed sperm storage tubules (SSTs) and allows for prolonged storage of semen, though the molecular mechanisms involved in semen preservation are not well understood. Little work has been done examining how function of the SSTs is impacted by insemination and by semen present in the SSTs. Methods: Transcriptome analysis was performed on isolated SSTs from turkey hens receiving no insemination (control), sham-insemination, or semen-insemination at three timepoints (D1, D30, and D90 post-insemination). Bioinformatic and functional annotation analyses were performed using CLC Genomics Workbench, Database for Annotation, Visualization, and Integrated Discovery (DAVID), and Ingenuity Pathway Analysis (IPA). Pairwise comparisons and k-medoids cluster analysis were utilized to decipher differential expression profiles in the treatment groups. Results: The SST transcriptome of the semen inseminated group exhibited the greatest differences within the group, with differences detectable for up to 90 days post insemination, while control and sham-inseminated groups were more similar. In the semen-inseminated samples, upregulation of pathways relating to classical and non-classical reproductive signaling, cytoskeletal remodeling, physiological parameters of the local UVJ environment, and cellular metabolism was observed. In the sham-inseminated samples, upregulation of immune pathways and non-reproductive endocrine hormones was observed. Discussion: This work provides insights into the molecular level changes of the SST in response to insemination as well as to the presence of semen. Results from this study may have direct implications on fertility rates as well as potential strategies for avian semen cryopreservation protocols.
Keywords: Turkey; artificial insemination; fertility; sperm storage tubule; transcriptome.
Copyright © 2024 Brady, Krasnec, Hanlon and Long.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Prolonged repeated inseminations trigger a local immune response and accelerate aging of the uterovaginal junction in turkey hens.Front Physiol. 2023 Nov 22;14:1275922. doi: 10.3389/fphys.2023.1275922. eCollection 2023. Front Physiol. 2023. PMID: 38074318 Free PMC article.
-
Transcriptome analysis of inseminated sperm storage tubules throughout the duration of fertility in the domestic turkey, Meleagris gallopavo.Poult Sci. 2022 Apr;101(4):101704. doi: 10.1016/j.psj.2022.101704. Epub 2022 Jan 10. Poult Sci. 2022. PMID: 35139440 Free PMC article.
-
Artificial insemination and laying cycle influence adaptive immune cell numbers and localization in the reproductive tract of turkey breeder hens.Poult Sci. 2024 Dec;103(12):104448. doi: 10.1016/j.psj.2024.104448. Epub 2024 Oct 28. Poult Sci. 2024. PMID: 39527869 Free PMC article.
-
Low dose insemination of mares using non-sorted and sex-sorted sperm.Anim Reprod Sci. 2001 Dec 3;68(3-4):279-89. doi: 10.1016/s0378-4320(01)00165-8. Anim Reprod Sci. 2001. PMID: 11744272 Review.
-
Sperm storage and transport following natural mating and artificial insemination.Poult Sci. 1993 May;72(5):923-8. doi: 10.3382/ps.0720923. Poult Sci. 1993. PMID: 8502613 Review.
Cited by
-
Fertility and Insemination Characteristics of Sperm Storage Tubules in Old Thai-Native Hens.Animals (Basel). 2024 Feb 23;14(5):694. doi: 10.3390/ani14050694. Animals (Basel). 2024. PMID: 38473079 Free PMC article.
References
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Bakst M. R. (1987). Anatomical basis of sperm-storage in the avian oviduct. Scanning Microsc. 1, 1257–1266. - PubMed
LinkOut - more resources
Full Text Sources

