This is a preprint.
Serum proteomics reveals APOE dependent and independent protein signatures in Alzheimer's disease
- PMID: 38260284
- PMCID: PMC10802738
- DOI: 10.21203/rs.3.rs-3706206/v1
Serum proteomics reveals APOE dependent and independent protein signatures in Alzheimer's disease
Update in
-
Serum proteomics reveal APOE-ε4-dependent and APOE-ε4-independent protein signatures in Alzheimer's disease.Nat Aging. 2024 Oct;4(10):1446-1464. doi: 10.1038/s43587-024-00693-1. Epub 2024 Aug 21. Nat Aging. 2024. PMID: 39169269 Free PMC article.
Abstract
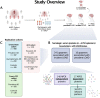
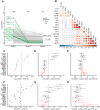
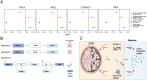
The current demand for early intervention, prevention, and treatment of late onset Alzheimer's disease (LOAD) warrants deeper understanding of the underlying molecular processes which could contribute to biomarker and drug target discovery. Utilizing high-throughput proteomic measurements in serum from a prospective population-based cohort of older adults (n = 5,294), we identified 303 unique proteins associated with incident LOAD (median follow-up 12.8 years). Over 40% of these proteins were associated with LOAD independently of APOE-ε4 carrier status. These proteins were implicated in neuronal processes and overlapped with protein signatures of LOAD in brain and cerebrospinal fluid. We found 17 proteins which LOAD-association was strongly dependent on APOE-ε4 carrier status. Most of them showed consistent associations with LOAD in cerebrospinal fluid and a third had brain-specific gene expression. Remarkably, four proteins in this group (TBCA, ARL2, S100A13 and IRF6) were downregulated by APOE-ε4 yet upregulated as a consequence of LOAD as determined in a bi-directional Mendelian randomization analysis, reflecting a potential response to the disease onset. Accordingly, the direct association of these proteins to LOAD was reversed upon APOE-ε4 genotype adjustment, a finding which we replicate in an external cohort (n = 719). Our findings provide an insight into the dysregulated pathways that may lead to the development and early detection of LOAD, including those both independent and dependent on APOE-ε4. Importantly, many of the LOAD-associated proteins we find in the circulation have been found to be expressed - and have a direct link with AD - in brain tissue. Thus, the proteins identified here, and their upstream modulating pathways, provide a new source of circulating biomarker and therapeutic target candidates for LOAD.
Keywords: APOE; Alzheimer’s disease; Brain; CSF; Cross-sectional study; Longitudinal study; Mendelian randomization; Network; Proteomics.
Conflict of interest statement
Competing interest declaration R.P., X.Q., N.F., L.L.J., A.P.O and J.J.L are employees and stockholders of Novartis. N.T.S and A.I.L are co-founders of Emtherapro. No other potential conflicts of interest relevant to this article were reported.
Figures




References
-
- Gatz M et al. (2006) Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63:168–174 - PubMed
-
- van Dyck CH et al. (2023) Lecanemab in Early Alzheimer’s Disease. N Engl J Med 388:9–21 - PubMed
-
- Mintun MA et al. (2021) Donanemab in Early Alzheimer’s Disease. N Engl J Med 384:1691–1704 - PubMed
Methods references
-
- Jørgensen L. M., El Kholy K., Damkjær K., Deis A. & Schroll M. »RAI« - Et internationalt system til vurdering af beboere på plejehjem. Ugeskr Laeger 159, 6371–6376 (1997). - PubMed
-
- Gudnason V S. J. S. L. H. S. S. G. Association of apolipoprotein E polymorphism with plasma levels of high density lipoprotein and lipoprotein(a), and effect of diet in healthy men and women. NUTRITION METABOLISM AND CARDIOVASCULAR DISEASES 3, 136–141 (1993).
-
- Levey A., Greene T., Kusek J. & Beck G. A simplified equation to predict glomerular filtration rate from serum creatinine. Journal of the American Society of Nephrology 11, 155A (2000).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

