This is a preprint.
Generative modeling of biological shapes and images using a probabilistic -shape sampler
- PMID: 38260340
- PMCID: PMC10802457
- DOI: 10.1101/2024.01.09.574919
Generative modeling of biological shapes and images using a probabilistic -shape sampler
Abstract
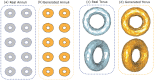
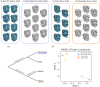
Understanding morphological variation is an important task in many areas of computational biology. Recent studies have focused on developing computational tools for the task of sub-image selection which aims at identifying structural features that best describe the variation between classes of shapes. A major part in assessing the utility of these approaches is to demonstrate their performance on both simulated and real datasets. However, when creating a model for shape statistics, real data can be difficult to access and the sample sizes for these data are often small due to them being expensive to collect. Meanwhile, the current landscape of generative models for shapes has been mostly limited to approaches that use black-box inference-making it difficult to systematically assess the power and calibration of sub-image models. In this paper, we introduce the -shape sampler: a probabilistic framework for generating realistic 2D and 3D shapes based on probability distributions which can be learned from real data. We demonstrate our framework using proof-of-concept examples and in two real applications in biology where we generate (i) 2D images of healthy and septic neutrophils and (ii) 3D computed tomography (CT) scans of primate mandibular molars. The -shape sampler R package is open-source and can be downloaded at https://github.com/lcrawlab/ashapesampler.
Figures



References
-
- Crawford Lorin, Monod Anthea, Chen Andrew X., Mukerhjee Sayan, and Rabadán Raúl. Predicting clinical outcomes in glioblastoma: An application of topological and functional data analysis. Journal of the American Statistical Association, 115:1139–1150, 2020. doi: 10.1080/01621459.2019.1671198. - DOI
-
- Boyer Doug M., Lipman Yaron, Clair Elizabeth, Puente Jesus, Patel Biren A., Funkhouser Thomas, Jernvall Jukka, and Ingrid Daubechies. Algorithms to automatically quantify the geometric similarity of anatomical surfaces. Proceedings of the National Academy of Sciences, 108(45):18221–18226, 2011. ISSN 0027-8424, 1091-6490. doi: 10.1073/pnas.1112822108. URL https://pnas.org/doi/full/10.1073/pnas.1112822108. - DOI - DOI - PMC - PubMed
-
- Theska Tobias, Sieriebriennikov Bogdan, Wighard Sara S, Werner Michael S, and Sommer Ralf J, Geometric morphometrics of microscopic animals as exemplified by model nematodes. Nature Protocols, pages 1–34, 2020. - PubMed
-
- Evans Kory M., Larouche Olivier, Watson Sara-Jane, Farina Stacy, Habegger María Laura, and Friedman Matt. Integration drives rapid phenotypic evolution in flatfishes. Proceedings of the National Academy of Sciences, 118(18):e2101330118, 2021. doi: 10.1073/pnas.2101330118. URL https://www.pnas.org/doi/abs/10.1073/pnas.2101330118. - DOI - DOI - PMC - PubMed
-
- Pincus Zachary and Theriot JA. Comparison of quantitative methods for cell-shape analysis. Journal of Microscopy, 227(2):140–156, 2007. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
