This is a preprint.
Alpha-synuclein regulates nucleolar DNA double-strand break repair in melanoma
- PMID: 38260370
- PMCID: PMC10802588
- DOI: 10.1101/2024.01.13.575526
Alpha-synuclein regulates nucleolar DNA double-strand break repair in melanoma
Update in
-
Alpha-synuclein regulates nucleolar DNA double-strand break repair in melanoma.Sci Adv. 2025 Apr 11;11(15):eadq2519. doi: 10.1126/sciadv.adq2519. Epub 2025 Apr 9. Sci Adv. 2025. PMID: 40203113 Free PMC article.
Abstract
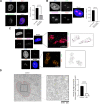
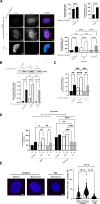
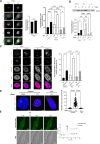
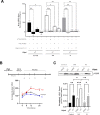
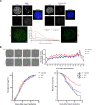
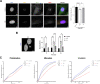
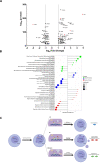
Although an increased risk of the skin cancer melanoma in people with Parkinson's Disease (PD) has been shown in multiple studies, the mechanisms involved are poorly understood, but increased expression of the PD-associated protein alpha-synuclein (αSyn) in melanoma cells may be important. Our previous work suggests that αSyn can facilitate DNA double-strand break (DSB) repair, promoting genomic stability. We now show that αSyn is preferentially enriched within the nucleolus in the SK-MEL28 melanoma cell line, where it colocalizes with DNA damage markers and DSBs. Inducing DSBs specifically within nucleolar ribosomal DNA (rDNA) increases αSyn levels near sites of damage. αSyn knockout increases DNA damage within the nucleolus at baseline, after specific rDNA DSB induction, and prolongs the rate of recovery from this induced damage. αSyn is important downstream of ATM signaling to facilitate 53BP1 recruitment to DSBs, reducing micronuclei formation and promoting cellular proliferation, migration, and invasion.
Conflict of interest statement
Potential Conflicts of Interest The authors declare no competing interests.
Figures

References
-
- Skibba JL, Pinckley J, Gilbert EF, Johnson RO. Multiple primary melanoma following administration of levodopa. Arch Pathol. Jun 1972;93(6):556–61. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
