This is a preprint.
D-type cyclins regulate DNA mismatch repair in the G1 and S phases of the cell cycle, maintaining genome stability
- PMID: 38260436
- PMCID: PMC10802603
- DOI: 10.1101/2024.01.12.575420
D-type cyclins regulate DNA mismatch repair in the G1 and S phases of the cell cycle, maintaining genome stability
Update in
-
CDK-independent role of D-type cyclins in regulating DNA mismatch repair.Mol Cell. 2024 Apr 4;84(7):1224-1242.e13. doi: 10.1016/j.molcel.2024.02.010. Epub 2024 Mar 7. Mol Cell. 2024. PMID: 38458201 Free PMC article.
Abstract
The large majority of oxidative DNA lesions occurring in the G1 phase of the cell cycle are repaired by base excision repair (BER) rather than mismatch repair (MMR) to avoid long resections that can lead to genomic instability and cell death. However, the molecular mechanisms dictating pathway choice between MMR and BER have remained unknown. Here, we show that, during G1, D-type cyclins are recruited to sites of oxidative DNA damage in a PCNA- and p21-dependent manner. D-type cyclins shield p21 from its two ubiquitin ligases CRL1SKP2 and CRL4CDT2 in a CDK4/6-independent manner. In turn, p21 competes through its PCNA-interacting protein degron with MMR components for their binding to PCNA. This inhibits MMR while not affecting BER. At the G1/S transition, the CRL4AMBRA1-dependent degradation of D-type cyclins renders p21 susceptible to proteolysis. These timely degradation events allow the proper binding of MMR proteins to PCNA, enabling the repair of DNA replication errors. Persistent expression of cyclin D1 during S-phase increases the mutational burden and promotes microsatellite instability. Thus, the expression of D-type cyclins inhibits MMR in G1, whereas their degradation is necessary for proper MMR function in S.
Conflict of interest statement
Declaration of interests PS has been a consultant for Novartis, Genovis, Guidepoint, The Planning Shop, ORIC Pharmaceuticals, Cedilla Therapeutics, Syros Pharmaceuticals, Exo Therapeutics, Curie Bio Operations, Exscientia, Ligature Therapeutics, Redesign Science, Blueprint and Merck; his laboratory receives research funding from Novartis. MP is a scientific cofounder of SEED Therapeutics; receives research funding from and is a shareholder in Kymera Therapeutics; and is a consultant for, a member of the scientific advisory board of, and has financial interests in CullGen, SEED Therapeutics, Triana Biomedicines, and Umbra Therapeutics; however, no research funds were received from these entities, and the findings presented in this manuscript were not discussed with any person in these companies. The other authors have no competing interests to declare.
Figures
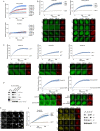
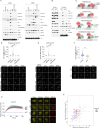





References
-
- Lukas J., Pagano M., Staskova Z., Draetta G., and Bartek J. (1994). Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumour cell lines. Oncogene 9, 707–718. - PubMed
-
- Zerjatke T., Gak I.A., Kirova D., Fuhrmann M., Daniel K., Gonciarz M., Muller D., Glauche I., and Mansfeld J. (2017). Quantitative Cell Cycle Analysis Based on an Endogenous All-in-One Reporter for Cell Tracking and Classification. Cell Rep 19, 1953–1966. 10.1016/j.celrep.2017.05.022. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
