This is a preprint.
Mechanosensitive genomic enhancers potentiate the cellular response to matrix stiffness
- PMID: 38260455
- PMCID: PMC10802421
- DOI: 10.1101/2024.01.10.574997
Mechanosensitive genomic enhancers potentiate the cellular response to matrix stiffness
Update in
-
Mechanosensitive genomic enhancers potentiate the cellular response to matrix stiffness.Science. 2025 Sep 25:eadl1988. doi: 10.1126/science.adl1988. Online ahead of print. Science. 2025. PMID: 40997217
Abstract
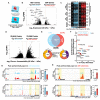
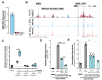
Epigenetic control of cellular transcription and phenotype is influenced by changes in the cellular microenvironment, yet how mechanical cues from these microenvironments precisely influence epigenetic state to regulate transcription remains largely unmapped. Here, we combine genome-wide epigenome profiling, epigenome editing, and phenotypic and single-cell RNA-seq CRISPR screening to identify a new class of genomic enhancers that responds to the mechanical microenvironment. These 'mechanoenhancers' could be active on either soft or stiff extracellular matrix contexts, and regulated transcription to influence critical cell functions including apoptosis, mechanotransduction, proliferation, and migration. Epigenetic editing of mechanoenhancers on rigid materials tuned gene expression to levels observed on softer materials, thereby reprogramming the cellular response to the mechanical microenvironment. These editing approaches may enable the precise alteration of mechanically-driven disease states.
Conflict of interest statement
Competing interests: C.A.G. is a co-founder of Tune Therapeutics and Locus Biosciences, and an advisor to Tune Therapeutics and Sarepta Therapeutics. C.A.G. is an inventor on patents and patent applications related to CRISPR epigenome editing. B.D.C. is an employee of Tune Therapeutics. The remaining authors have no conflicts of interest to declare.
Figures



References
-
- Jones D. L., Wagers A. J., No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 9, 11–21 (2008). - PubMed
-
- Simeonov D. R., Gowen B. G., Boontanrart M., Roth T. L., Gagnon J. D., Mumbach M. R., Satpathy A. T., Lee Y., Bray N. L., Chan A. Y., Lituiev D. S., Nguyen M. L., Gate R. E., Subramaniam M., Li Z., Woo J. M., Mitros T., Ray G. J., Curie G. L., Naddaf N., Chu J. S., Ma H., Boyer E., Van Gool F., Huang H., Liu R., Tobin V. R., Schumann K., Daly M. J., Farh K. K., Ansel K. M., Ye C. J., Greenleaf W. J., Anderson M. S., Bluestone J. A., Chang H. Y., Corn J. E., Marson A., Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 549, 111–115 (2017). - PMC - PubMed
-
- Korkmaz G., Lopes R., Ugalde A. P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R., Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 34, 192–198 (2016). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
