This is a preprint.
Lupus-associated innate receptors drive extrafollicular evolution of autoreactive B cells
- PMID: 38260501
- PMCID: PMC10802414
- DOI: 10.1101/2024.01.09.574739
Lupus-associated innate receptors drive extrafollicular evolution of autoreactive B cells
Abstract
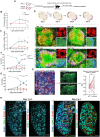
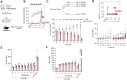
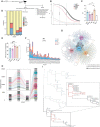
In systemic lupus erythematosus, recent findings highlight the extrafollicular (EF) pathway as prominent origin of autoantibody-secreting cells (ASCs). CD21loCD11c+ B cells, associated with aging, infection, and autoimmunity, are contributors to autoreactive EF ASCs but have an obscure developmental trajectory. To study EF kinetics of autoreactive B cell in tissue, we adoptively transferred WT and gene knockout B cell populations into the 564Igi mice - an autoreactive host enriched with autoantigens and T cell help. Time-stamped analyses revealed TLR7 dependence in early escape of peripheral B cell tolerance and establishment of a pre-ASC division program. We propose CD21lo cells as precursors to EF ASCs due to their elevated TLR7 sensitivity and proliferative nature. Blocking receptor function reversed CD21 loss and reduced effector cell generation, portraying CD21 as a differentiation initiator and a possible target for autoreactive B cell suppression. Repertoire analysis further delineated proto-autoreactive B cell selection and receptor evolution toward self-reactivity. This work elucidates receptor and clonal dynamics in EF development of autoreactive B cells, and establishes modular, native systems to probe mechanisms of autoreactivity.
Figures




Similar articles
-
The extrafollicular response is sufficient to drive initiation of autoimmunity and early disease hallmarks of lupus.Front Immunol. 2022 Dec 14;13:1021370. doi: 10.3389/fimmu.2022.1021370. eCollection 2022. Front Immunol. 2022. PMID: 36591222 Free PMC article.
-
Facultative role for T cells in extrafollicular Toll-like receptor-dependent autoreactive B-cell responses in vivo.Proc Natl Acad Sci U S A. 2011 May 10;108(19):7932-7. doi: 10.1073/pnas.1018571108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518858 Free PMC article.
-
Extrafollicular responses in humans and SLE.Immunol Rev. 2019 Mar;288(1):136-148. doi: 10.1111/imr.12741. Immunol Rev. 2019. PMID: 30874345 Free PMC article. Review.
-
Multiple checkpoint breach of B cell tolerance in Rasgrp1-deficient mice.J Immunol. 2013 Oct 1;191(7):3605-13. doi: 10.4049/jimmunol.1202892. Epub 2013 Aug 30. J Immunol. 2013. PMID: 23997211
-
Mechanisms of autoantibody production in systemic lupus erythematosus.Front Immunol. 2015 May 13;6:228. doi: 10.3389/fimmu.2015.00228. eCollection 2015. Front Immunol. 2015. PMID: 26029213 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
