This is a preprint.
Mitochondrial membrane potential regulates nuclear DNA methylation and gene expression through phospholipid remodeling
- PMID: 38260521
- PMCID: PMC10802563
- DOI: 10.1101/2024.01.12.575075
Mitochondrial membrane potential regulates nuclear DNA methylation and gene expression through phospholipid remodeling
Update in
-
Mitochondrial membrane hyperpolarization modulates nuclear DNA methylation and gene expression through phospholipid remodeling.Nat Commun. 2025 Apr 29;16(1):4029. doi: 10.1038/s41467-025-59427-5. Nat Commun. 2025. PMID: 40301431 Free PMC article.
Abstract
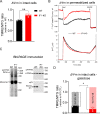
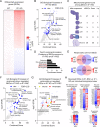
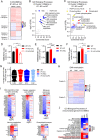
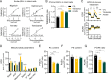
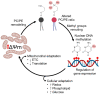
Maintenance of the mitochondrial inner membrane potential (ΔΨM) is critical for many aspects of mitochondrial function, including mitochondrial protein import and ion homeostasis. While ΔΨM loss and its consequences are well studied, little is known about the effects of increased ΔΨM. In this study, we used cells deleted of ATPIF1, a natural inhibitor of the hydrolytic activity of the ATP synthase, as a genetic model of mitochondrial hyperpolarization. Our data show that chronic ΔΨM increase leads to nuclear DNA hypermethylation, regulating transcription of mitochondria, carbohydrate and lipid metabolism genes. Surprisingly, remodeling of phospholipids, but not metabolites or redox changes, mechanistically links the ΔΨM to the epigenome. These changes were also observed upon chemical exposures and reversed by decreasing the ΔΨM, highlighting them as hallmark adaptations to chronic mitochondrial hyperpolarization. Our results reveal the ΔΨM as the upstream signal conveying the mitochondrial status to the epigenome to regulate cellular biology, providing a new framework for how mitochondria can influence health outcomes in the absence of canonical dysfunction.
Keywords: DNA hypermethylation; Mitochondria; epigenome; gene expression; mitochondrial membrane potential; phospholipid rewiring.
Conflict of interest statement
Declaration of Interests The authors declare no conflict of interest.
Figures

References
-
- Di Lisa F., Blank P.S., Collona R., Gambassi G., Silverman H.S., Stern M.S., and Hansford R.G. (1995). Mitochondrial membrane potential in single living adult rat cardiac myocytes exposed to anoxia or metabolic inhibition. The Journal of Physiology 486, 1–13. 10.1113/jphysiol.1995.sp020786. - DOI - PMC - PubMed
-
- Pak O., Sommer N., Hoeres T., Bakr A., Waisbrod S., Sydykov A., Haag D., Esfandiary A., Kojonazarov B., Veit F., et al. (2013). Mitochondrial hyperpolarization in pulmonary vascular remodeling. Mitochondrial uncoupling protein deficiency as disease model. Am J Respir Cell Mol Biol 49, 358–367. 10.1165/rcmb.2012-0361OC. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
