Diazotrophic abundance and community structure associated with three meadow plants on the Qinghai-Tibet Plateau
- PMID: 38260880
- PMCID: PMC10801153
- DOI: 10.3389/fmicb.2023.1292860
Diazotrophic abundance and community structure associated with three meadow plants on the Qinghai-Tibet Plateau
Abstract
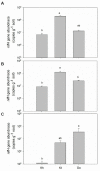
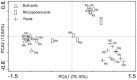
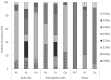
Symbiotic diazotrophs form associations with legumes and substantially fix nitrogen into soils. However, grasslands on the Qinghai-Tibet Plateau are dominated by non-legume plants, such as Kobresia tibetica. Herein, we investigated the diazotrophic abundance, composition, and community structure in the soils and roots of three plants, non-legume K. tibetica and Kobresia humilis and the legume Oxytropis ochrocephala, using molecular methods targeting nifH gene. Diazotrophs were abundantly observed in both bulk and rhizosphere soils, as well as in roots of all three plants, but their abundance varied with plant type and soil. In both bulk and rhizosphere soils, K. tibetica showed the highest diazotroph abundance, whereas K. humilis had the lowest. In roots, O. ochrocephala and K. humilis showed the highest and the lowest diazotroph abundance, respectively. The bulk and rhizosphere soils exhibited similar diazotrophic community structure in both O. ochrocephala and K. tibetica, but were substantially distinct from the roots in both plants. Interestingly, the root diazotrophic community structures in legume O. ochrocephala and non-legume K. tibetica were similar. Diazotrophs in bulk and rhizosphere soils were more diverse than those in the roots of three plants. Rhizosphere soils of K. humilis were dominated by Actinobacteria, while rhizosphere soils and roots of K. tibetica were dominated by Verrumicrobia and Proteobacteria. The O. ochrocephala root diazotrophs were dominated by Alphaproteobacteria. These findings indicate that free-living diazotrophs abundantly and diversely occur in grassland soils dominated by non-legume plants, suggesting that these diazotrophs may play important roles in fixing nitrogen into soils on the plateau.
Keywords: Qinghai-Tibet Plateau; biological nitrogen fixation; diazotrophs; grassland ecosystems; nifH gene; non-leguminous plants.
Copyright © 2024 Nshimiyimana, Zhao, Wang and Kong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Altitudinal niches of symbiotic, associative and free-living diazotrophs driven by soil moisture and temperature in the alpine meadow on the Tibetan Plateau.Environ Res. 2022 Aug;211:113033. doi: 10.1016/j.envres.2022.113033. Epub 2022 Mar 8. Environ Res. 2022. PMID: 35276191
-
Distribution of rhizosphere fungi of Kobresia humilis on the Qinghai-Tibet Plateau.PeerJ. 2024 Feb 20;12:e16620. doi: 10.7717/peerj.16620. eCollection 2024. PeerJ. 2024. PMID: 38406296 Free PMC article.
-
Autotrophic and symbiotic diazotrophs dominate nitrogen-fixing communities in Tibetan grassland soils.Sci Total Environ. 2018 Oct 15;639:997-1006. doi: 10.1016/j.scitotenv.2018.05.238. Epub 2018 May 26. Sci Total Environ. 2018. PMID: 29929338
-
Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants.Plants (Basel). 2019 Oct 11;8(10):408. doi: 10.3390/plants8100408. Plants (Basel). 2019. PMID: 31614562 Free PMC article. Review.
-
Nitrogen Fixation in Cereals.Front Microbiol. 2018 Aug 9;9:1794. doi: 10.3389/fmicb.2018.01794. eCollection 2018. Front Microbiol. 2018. PMID: 30140262 Free PMC article. Review.
References
-
- Barros B. G. D. F., Freitas A. D. S. D., Tabosa J. N., Lyra M. D. C. C. P. D., Mergulhão A. C. D. E. S., Silva A. F. D., et al. . (2020). Biological nitrogen fixation in field-grown sorghum under different edaphoclimatic conditions is confirmed by N isotopic signatures. Nutr. Cycl. Agroecosystems. 117, 93–101. doi: 10.1007/s10705-020-10051-3 - DOI
-
- Bohlool B. B., Ladha J. K., Garrity D. P., George T. (1992). Biological nitrogen fixation for sustainable agriculture: a perspective. Plant Soil 141, 1–11. doi: 10.1007/BF00011307 - DOI
LinkOut - more resources
Full Text Sources

